I just finished reading two really great books by Nick Lane, a biochemist at the University College London - Life Ascending: The Ten Great Inventions of Evolution (2010) and The Vital Question: Energy, Evolution, and the Origins of Complex Life (2015). I am certainly not alone on this. Bill Gates has also recommended that we all read the The Vital Question. Now in this posting, I would like to focus on Nick Lane's vital question of how living things
stumbled upon complexity on this planet and relate his findings to how software converged upon the very same solution through a similar process. Recall that one of the fundamental findings of softwarephysics is that living things and software are both forms of self-replicating information and that both have converged upon similar solutions to combat the second law of thermodynamics in a highly nonlinear Universe. For biologists, the value of softwarephysics is that software has been evolving about 100 million times faster than living things over the past 76 years, or 2.4 billion seconds, ever since Konrad Zuse first cranked up his Z3 computer in May of 1941, and the evolution of software over that period of time is the only history of a form of self-replicating information that has actually been recorded by human history. In fact, the evolutionary history of software has all occurred within a single human lifetime, and many of those humans are still alive today to testify as to what actually had happened, something that those working on the origin of life on the Earth and its early evolution can only try to imagine. Again, in softwarephysics, we define self-replicating information as:
Self-Replicating Information – Information that persists through time by making copies of itself or by enlisting the support of other things to ensure that copies of itself are made.
The Characteristics of Self-Replicating Information
All forms of self-replicating information have some common characteristics:
1. All self-replicating information evolves over time through the Darwinian processes of inheritance, innovation and natural selection, which endows self-replicating information with one telling characteristic – the ability to survive in a Universe dominated by the second law of thermodynamics and nonlinearity.
2. All self-replicating information begins spontaneously as a parasitic mutation that obtains energy, information and sometimes matter from a host.
3. With time, the parasitic self-replicating information takes on a symbiotic relationship with its host.
4. Eventually, the self-replicating information becomes one with its host through the symbiotic integration of the host and the self-replicating information.
5. Ultimately, the self-replicating information replaces its host as the dominant form of self-replicating information.
6. Most hosts are also forms of self-replicating information.
7. All self-replicating information has to be a little bit nasty in order to survive.
8. The defining characteristic of self-replicating information is the ability of self-replicating information to change the boundary conditions of its utility phase space in new and unpredictable ways by means of exapting current functions into new uses that change the size and shape of its particular utility phase space. See Enablement - the Definitive Characteristic of Living Things for more on this last characteristic.
So far we have seen 5 waves of self-replicating information sweep across the Earth, with each wave greatly reworking the surface and near subsurface of the planet as it came to predominance:
1. Self-replicating autocatalytic metabolic pathways of organic molecules
2. RNA
3. DNA
4. Memes
5. Software
Some object to the idea of memes and software being forms of self-replicating information because currently, both are products of the human mind. But I think that objection stems from the fact that most people simply do not consider themselves to be a part of the natural world. Instead, most people consciously or subconsciously consider themselves to be a supernatural and immaterial spirit that is temporarily haunting a carbon-based body. Now, in order for evolution to take place, we need all three Darwinian processes at work – inheritance, innovation and natural selection. And that is the case for all forms of self-replicating information, including carbon-based life, memes and software. Currently, software is being written and maintained by human programmers, but that will likely change in the next 10 – 50 years when the Software Singularity occurs and AI software will be able to write and maintain software better than a human programmer. Even so, one must realize that human programmers are also just machines with a very complicated and huge neural network of neurons that have been trained with very advanced Deep Learning techniques to code software. Nobody learned how to code software sitting alone in a dark room. All programmers inherited the memes for writing software from teachers, books, other programmers or by looking at the code of others. Also, all forms of selection are "natural" unless they are made by supernatural means. So a programmer pursuing bug-free software by means of trial and error is no different than a cheetah deciding upon which gazelle in a herd to pursue. Software is now rapidly becoming the dominant form of self-replicating information on the planet and is having a major impact on mankind as it comes to predominance. For more on that see: A Brief History of Self-Replicating Information.
The Strong Connection Between Information Flows and Energy Flows
In biology today there seems to be a disturbing ongoing battle between bioinformatics, the study of information flows in biology, and bioenergetics, the study of energy flows in biology. Because of the great triumphs that bioinformatics has made in the 20th and 21st centuries, and the huge amount of attention it has garnered in the public domain, bioenergetics has, unfortunately, been undeservedly relegated somewhat to a backburner in biology, relative to the esteemed position it once held earlier in the 20th century when people were actively working on things like the Krebs cycle. This is indeed unfortunate because information flows and energy flows in our Universe are intimately connected. I think that much of the confusion arises from the rather ambiguous definition of the term "information" that is in common use today. In The Demon of Software I explained that in softwarephysics we exclusively use Leon Brillouin’s concept of information as a form of negative entropy. In that view, in order to create some useful information, like a chunk of functional software, one must first degrade some high-grade form of energy, like electrical energy, into low-grade heat energy. Thanks to the first law of thermodynamics, no energy is lost in the process of creating useful information, but thanks to the second law of thermodynamics, we must always convert some high-grade form of energy into an equal amount of low-grade heat energy in order to make it all happen. Similarly, using Leon Brillouin’s concept of information, the bioenergetics of a very efficient biochemical metabolic pathway also becomes a form of useful information, like a chunk of functional software. In contrast, electrical engineers and those in bioinformatics, usually think of information in terms of Claude Shannon's view that information is the amount of "surprise" in a message. That is because they are primarily concerned with the DNA sequences of a species and comparing the DNA sequences of proteins between species. For more on that please see: Some More Information About Information. Now Nick Lane is definitely a member of the bioenergetics camp, so he might not be so pleased with softwarephysics, and its seeming obsession with the characteristics of self-replicating information, but that might be because he is most familiar with the people in bioinformatics who primarily use Claude Shannon's view that information is the amount of "surprise" in a message. However, I think that Nick Lane would be quite comfortable with softwarephysics' use of Leon Brillouin’s concept of information as a form of negative entropy, that can only be created by a heat engine that degrades high-grade energy into low-grade heat energy, because in that view, an efficient metabolic pathway that has been honed by natural selection is a form of negative entropy, and is therefore, also a useful form of information too - something that both bioinformatics and bioenergetics can easily share. In that view, bioinformatics and bioenergetics go hand-in-hand and become essentially one.
In IT we also deal with both formulations for the concept of information. We deal with large amounts of data, like customer and product data, that is then processed by software algorithms. The customer and product data are very much like DNA sequences, where Claude Shannon's view of information as the amount of surprise in a message makes sense, but the processing software algorithms are more like bioenergetic metabolic pathways, where Leon Brillouin’s concept of information as a form of negative entropy is more appropriate.
The Great Mystery of Complexity in Biology
In the The Vital Question, Nick Lane addresses the greatest mystery of them all in biology - why there is complex life on the planet and why all of that complex life is based upon exactly the same identical cellular architecture. Now in discussing complex life on the planet, most people are usually talking about things like lions, tigers and bears - "oh my!". But Nick Lane is really talking about the very complex architecture of eukaryotic cells, as opposed to the much simpler architecture of prokaryotic cells, because lions, tigers and bears, and all of the other "higher" forms of life that we are familiar with are simply made of aggregations of eukaryotic cells. Even the simple yeasts that make our breads, and get us drunk, are very complex eukaryotic cells. The troubling thing is that only an expert could tell the difference between a yeast eukaryotic cell and a human eukaryotic cell because they are so similar, while any school child could easily tell the difference between the microscopic images of a prokaryotic bacterial cell and a eukaryotic yeast cell - see Figure 1.
Figure 1 – The prokaryotic cell architecture of the bacteria and archaea is very simple and designed for rapid replication. Prokaryotic cells do not have a nucleus enclosing their DNA. Eukaryotic cells, on the other hand, store their DNA on chromosomes that are isolated in a cellular nucleus. Eukaryotic cells also have a very complex internal structure with a large number of organelles, or subroutine functions, that compartmentalize the functions of life within the eukaryotic cells.
Prokaryotic cells essentially consist of a tough outer cell wall enclosing an inner cell membrane and contain a minimum of internal structure. The cell membrane is composed of phospholipids and proteins. The DNA within prokaryotic cells generally floats freely as a large loop of DNA, and their ribosomes, used to help translate mRNA into proteins, float freely within the entire cell as well. The ribosomes in prokaryotic cells are not attached to membranes like they are in eukaryotic cells which have membranes called the rough endoplasmic reticulum for that purpose. The chief advantage of prokaryotic cells is their simple design and the ability to thrive and rapidly reproduce even in very challenging environments, like little AK-47s that still manage to work in environments where modern tanks will fail. Eukaryotic cells, on the other hand, are found in the bodies of all complex organisms, from single-celled yeasts to you and me, and they divide up cell functions amongst a collection of organelles (functional subroutines), such as mitochondria, chloroplasts, Golgi bodies, and the endoplasmic reticulum.
Recall that we now know that there actually are three forms of life on this planet, as first described by Carl Woese in 1977 at my old Alma Mater the University of Illinois - the Bacteria, the Archea and the Eucarya. The Bacteria and the Archea both use the simple prokaryotic cell architecture, while the Eucarya use the much more complicated eukaryotic cell architecture. Figure 2 depicts Carl Woese's rewrite of Darwin's famous tree of life and shows that complex forms of life, like you and me, that are based upon cells using the eukaryotic cell architecture, actually spun off from the archaea and not the bacteria. Now archaea and bacteria look identical under a microscope, and that is the reason why at first we thought they were all just bacteria for hundreds of years. But in the 1970s Carl Woese discovered that the ribosomes used to transcribe mRNA into proteins were different between certain microorganisms that had all been previously lumped together as "bacteria". Carl Woese determined that the lumped together "bacteria" really consisted of two entirely different forms of life - the bacteria and the archaea - see Figure 2. The bacteria and archaea both have cell walls, but use slightly different organic molecules to build them. Some archaea, known as the extremophiles that live in harsh conditions also wrap their DNA around stabilizing histone proteins. Eukaryotes also wrap their DNA around histone proteins to form chromatin and chromosomes - for more on that see: An IT Perspective on the Origin of Chromatin, Chromosomes and Cancer. For that reason, and other biochemical reactions that the archaea and eukaryotes both share, we now think that the eukaryotes split off from the archaea and not the bacteria.
Figure 2 – In 1977 Carl Woese developed a new tree of life consisting of the Bacteria, the Archea and the Eucarya. The Bacteria and Archea use a simple prokaryotic cell architecture, while the Eucarya use the much more complicated eukaryotic cell structure.
The other thing about eukaryotic cells, as opposed to prokaryotic cells, is that eukaryotic cells are HUGE! They are like 15,000 times larger by volume than prokaryotic cells! See Figure 3 for a true-scale comparison of the two. The usual depiction of the differences between prokaryotic cells and eukaryotic cells, like that of Figure 2, reminds me very much of the very distorted picture of the Solar System that I grew up with. Unfortunately, most schoolchildren are frequently presented with a highly distorted depiction of our Solar System, in which the planets are much larger than they should be, and the orbits of the planets are much smaller than they should be too, relative to the size of the Sun, and unfortunately, this glaring distortion of the facts unknowingly haunts them for the rest of their lives! In such depictions, the size of the Sun and the orbits of the planets, and the sizes of the planets is highly distorted, as is depicted in Figure 4.
Figure 3 – Not only are eukaryotic cells much more complicated than prokaryotic cells, they are also HUGE!
Figure 4 – Schoolchildren are frequently presented with a highly distorted depiction of our Solar System, in which the planets are much larger than they should be, and the orbits of the planets are much smaller than they should be too.
Figure 5 – Above is an accurate depiction of the relative sizes of the planets compared to the Sun. If the Sun were the size of a standard basketball (9.39 inches or 0.239 meters in diameter), then the Earth would be a little smaller than a peppercorn (0.086 inches in diameter) at a distance of 84.13 feet (25.69 meters) and Jupiter would be at a distance of 437.87 feet (133.73 meters) from the Sun.
The Great Mystery of biology is that there are no examples of intermediate forms between the simple prokaryotic cell architecture and the very large and very complex eukaryotic cell architecture. Darwinian thought is based upon the evolution of life by means of inheritance and innovation honed by natural selection, and therefore, should lead to a continuous path through Design Space of intermediate forms between the simple prokaryotic cell architecture and the much larger and more complex eukaryotic cell architecture, but none is to be found in nature. Frequently, these intermediate forms are hard to find in the fossil record because many times the evolution from one form to another is very rapid in geological terms, and therefore, very few intermediate forms are left behind in the fossil record. But in recent decades we have indeed found many fossils of intermediate forms in the fossil record. Fortunately, we are not limited solely to the fossil record because many times nature leaves behind a trail of species that are stuck in intermediate forms as they converge upon a similar solution to a given problem.
The classic example of this is the evolution of the camera-like eye. Many of those in opposition to Darwinian thought frequently bring up the complex human camera-like eye as an example of something that could not possibly evolve by means of small incremental changes arising from inheritance and innovation honed by natural selection. They argue that 1% of a complex camera-like eye is of no value without the other 99% of the complex components. But that is not true. A 1% eye in the form of a light-patch of neurons that can be excited by incoming photons is certainly much better than being totally blind because it allows organisms to sense the shadow of a potential predator and then to get out of the way. Visible photons from the Sun have an energy of a few electron volts, and thus, could easily trigger chemical reactions that also only require a few electron volts of energy within some already existing neurons that initially evolved for an entirely different purpose. Thus, such neurons could easily become exapted into serving as the photoreceptors of a light-patch of an organism. Once a light-patch forms, a camera-like eye will soon follow. For example, a slight depression of the light-patch offers some degree of ability to detect the direction from which the incoming photons are coming, providing some directional information on how to avoid predators or to locate prey. As the depression of the light-patch deepens through small incremental changes, this ability to sense the direction of incoming photons also incrementally improves too. As the depression starts to become a nearly spherical pit with a small opening, it starts to begin to behave like a pinhole camera, forming a very dim blurry image back on the photoreceptors. Similarly, if the original skin over the light-patch begins to thicken into a lens-like shape, it will help to protect the photoreceptors on the back wall of the incipient eye, and it will also help to focus and sharpen the image formed by the incoming photons. Figure 8
shows that we can actually catch various species of organisms in the intermediate stages of a light-patch becoming a camera-like eye.
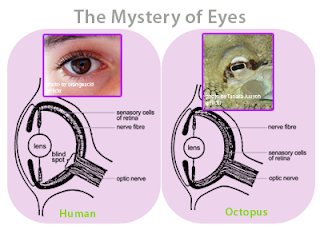
Figure 6 - The eye of a human and the eye of an octopus are nearly identical in structure, but evolved totally independently of each other. As Daniel Dennett pointed out, there are only a certain number of Good Tricks in Design Space and natural selection will drive different lines of descent towards them.
Figure 7 – Computer simulations reveal how a camera-like eye can easily evolve from a simple-light sensitive spot on the skin.
Figure 8 – We can actually see this evolutionary history unfold in the evolution of the camera-like eye by examining modern-day mollusks such as the octopus.
The biggest mystery in biology is that it seems as though the very large and very complex architecture of eukaryotic cells just seems to have suddenly popped into existence all by itself about 2 billion years ago, with no intermediates left behind between the simple prokaryotic cell architecture and the complex eukaryotic cell architecture. We simply now do not see any prokaryotic cells caught in the act of being at an intermediate stage on the way to becoming full-fledged eukaryotic cells. There are some creatures called archezoa that were once thought to be the "missing link" between the prokaryotes and the eukaryotes, but it now turns out that they are really just dumbed-down eukaryotic cells that lost some of the features of eukaryotic cells. But the archezoa do help to demonstrate that some intermediate forms could be viable, so why do we not see any?
Nick Lane maintains that there must be some extremely inhibitive limiting factor that has generally prevented prokaryotic cells from advancing on to become large complicated eukaryotic cells over the last 4.0 billion years, and also that some very rare event must have taken place about 2 billion years ago to breach that inhibitive wall, but never again. Nick Lane thinks that the limiting factor was a lack of free energy. Nick Lane goes on to explain that eukaryotic cells have access to about 5,000 times as much free energy as do prokaryotic cells, thanks to the work of several hundred enslaved mitochondria in each eukaryotic cell. Compared to the simple, but extremely energy-poor prokaryotic cells, eukaryotic cells are the super-rich in biology, with a tremendous abundance of free energy to build outlandish palatial estates, and that is what allowed prokaryotic archaeon cells to evolve into the much more complex eukaryotic cells of today.
Figure 9 – The juxtaposition of extreme poverty next to extreme wealth in Kenya, is a metaphor for the extreme energy poverty of prokaryotic cells, compared to the extreme energy abundance of the eukaryotic cells that have 5,000 times as much free energy to work with. With large amounts of free energy, it is possible to build the palatial estates of the eukaryotic cell architecture. Remember, in thermodynamics energy is the ability to do work, while in economics, money is the ability not to do work.
Recall that in the Endosymbiosis theory of Lynn Margulis, the mitochondria were originally parasitic bacteria that once invaded archaeon prokaryotic cells and took up residence. Certain of those ancient archaeon prokaryotic cells, with their internal bacterial mitochondrial parasites, were then able to survive the parasitic bacterial onslaught, and later, went on to form a strong parasitic/symbiotic relationship with them, like all forms of self-replicating information tend to do. The reason researchers think this is what happened is because mitochondria have their own DNA and that DNA is stored as a loose loop like bacteria store their DNA. Also, when a eukaryotic cell divides, hundreds of mitochondria first self-replicate, like a bacterial infection is want to do, just before the eukaryotic cell divides and half of the mitochondria are then passed on to each daughter cell like a bacterial infection does to self-replicate.
Figure 10 shows what the mitochondria do to power eukaryotic cells. Essentially they are friendly little bacterial parasites residing in eukaryotic cells, and the mitochondria contain a great deal of internal membrane surface area. Along these internal membranes is an electron transport chain that acts much like an energy delivery staircase. Willing electrons from the eukaryotic host cell are to be found on the organic molecules generated by the Krebs cycle within each mitochondrion, and those electrons then bounce down the electron transport chain, like a ball bouncing down a staircase. Each time the ball or electron bounces, it pumps an H+ proton to the outside of an internal membrane. Now an H+ proton is simply a hydrogen atom stripped of its electron, and the hydrogen atoms come from the NADH molecules produced by the Krebs cycle in the mitochondria. This causes a net positive charge to accumulate on the outside of the internal membrane and a net negative charge to form along the inside of the internal membrane, like a biological capacitor with the electric field strength of a lightning stroke, and this vast potential difference can be used to do work. For example, a number of H+ protons can drop back through a gate to synthesize ATP from ADP, and that ATP can be later used to fuel a biochemical process. Now it turns out that all living things use this same trick of pumping H+ protons across a membrane to later be used to do some useful biochemical work. It's a great way to store energy for later use because you can pump up small amounts of H+ protons and then later spend a large number of them on an expensive item, like saving up a few dollars from each paycheck so that you can later put down a down payment for a new car. In fact, Nick Lane begins the The Vital Question by explaining how alkaline hydrothermal vents are now our best bet for finding the original cradle of life on this planet. This is done in an excellent, and highly accessible manner, that would make reading The Vital Question well worth it for that alone. Alkaline hydrothermal vents provide a porous matrix that is infused with alkaline pore fluids, that are at a nice toasty temperature suitable for the manufacture of organic molecules, and which are produced by the serpentinization of the mineral olivine into the mineral serpentenite. These warm alkaline pore fluids also contain a great deal of dissolved hydrogen H2 gas too. The alkaline hydrothermal vent structures are sitting in acidic seawater containing a great deal of dissolved carbon dioxide CO2. The combination of all of this could allow for the formation of H+ proton gradients to form across inorganic iron sulfide FeS "membranes". That stored energy could then be used to make organic molecules out of H2 and CO2 - for more on that see: An IT Perspective on the Transition From Geochemistry to Biochemistry and Beyond.
Figure 10 – Mitochondria are little parasitic bacteria that at one time invaded some prokaryotic archaeon cells about 2 billion years ago, and went on to form a strong parasitic/symbiotic relationship with their archaeon hosts. Mitochondria have their own genes stored on bacterial DNA in a large loop, just like all other bacteria. Each eukaryotic cell contains several hundred mitochondria, which self-replicate before the eukaryotic cell divides. Half of the mitochondria go into each daughter cell after a division of the eukaryotic cell. The eukaryotic host cell provides the mitochondria with a source of food, and the mitochondria metabolize that food using the Krebs cycle and an electron transport chain to pump H+ protons uphill to the outside of their internal membranes. As the H+ protons fall back down they release stored energy to turn ADP into ATP for later use as a fuel.
Living Together Is Never Easy
Nick Lane then goes on to explain that living together is never easy. Sure, these early prokaryotic archaeon cells may have initially learned to live with their new energy-rich parasitic bacterial symbiotes, like an old man taking on a new trophy-wife, but there are always problems in cohabitating, like a number of new and unwelcome freeloading brothers-in-law taking up residence too. For example, bacteria carry with them parasitic segments of DNA called "mobile group II self-splicing introns". These are segments of parasitic DNA that are just trying to self-replicate, like all other forms of self-replicating information. These bacterial introns snip themselves out of the mRNA that is copied from the bacterial DNA and then form an active complex of reverse transcriptase that reinserts the intron DNA back into the bacterial DNA loop between genes to complete the self-replication process. These bacterial introns are rather mild parasites that just slow down the replication time for the main bacterial DNA loop, and also waste a bit of energy and material as they self-replicate. A typical bacterial cell with 4,000 genes might have about 30 of these introns, so over time, the bacteria just managed to learn to live with these pesky introns somehow. The bacteria that could not keep their introns under control simply died out, and so the bacteria infected with tons of parasitic introns were simply weeded out because they could not compete with the more healthy bacteria.
Now for the eukaryotes, the situation is completely different. Eukaryotes have tens of thousands of introns buried in their 20,000+ genes, and these introns are located right in the middle of the eukaryotic genes!
Figure 11 – Eukaryotic genes consist of a number of stretches of DNA, called exons, that code for protein synthesis, and a number of stretches of DNA, called introns, that do not code for protein synthesis. Unlike the bacteria, eukaryotic introns are not between the genes, they are right in the middle of the genes, so they must be spliced out of the transcribed mRNA within the eukaryotic nuclear membrane by molecular machines called spliceosomes before they exit a nuclear poor and become translated by ribosomes into a protein.
These introns must be spliced out of the transcribed mRNA by cellular machines called spliceosomes before the mRNA, transcribed from the genetic DNA, can be translated into a sequence of amino acids to form a protein. If the eukaryotic introns were not spliced out of the mRNA prior to translation by ribosomes, incorrect polypeptide chains of amino acids would form, creating proteins that simply would not work. This is a messy, but necessary process. Now the strange thing is that we share hundreds of genes with other forms of life, like trees, because we both evolved from some common ancestor, and we both still need some proteins that essentially perform the same biochemical functions. The weird thing is that these shared genes all have their introns in exactly the same location within these shared genes. This means that the parasitic introns must have been introduced into the DNA of the eukaryotic cell architecture very early, and that is why trees and humans have the introns located in the same spots on shared genes. The theory goes that when the parasitic mitochondria first took up residence in prokaryotic archaeon cells, like all parasites, they tried to take as much advantage of their hosts as possible, without killing the hosts outright. Now copying the several million base pairs in the 5,000 genes of a bacterial cell is the limiting time factor in self-replicating bacteria and requires at least 20 minutes to do so. Consequently, the mitochondrial invaders began to move most of their genes to the DNA loop of their hosts, and let their energy-rich archaeon hosts produce the proteins associated with those transplanted genes for them. However, the mitochondrial invaders wisely kept all of the genes necessary for metabolism locally on their own mitochondrial DNA loop because that allowed them to quickly get their hands on the metabolic proteins they needed without putting in a back order back on the main host DNA loop. After all, their energy-rich archaeon hosts now had several hundred mitochondria constantly pumping out the necessary ATP to make proteins, so why not let their hosts provide most of the proteins that were not essential to metabolism. This was the beginning of the compartmentalization of function within the hosts and was the beginning of a division of labor that produced a symbiotic advantage for both the archaeon hosts and their mitochondrial invaders.
But there was one problem with transplanting the mitochondrial genes to their hosts' DNA loop. The parasitic bacterial introns from the mitochondria tagged along as well, and those transplanted parasitic bacterial introns could now run wild because their energy-rich hosts could now afford the luxury of supporting tons of parasitic DNA, like a number of freeloading brothers-in-law that came along with your latest trophy wife - at least up to a point. Indeed, probably most archaeon hosts died from the onslaught of tons of parasitic mitochondrial introns clogging up their critical genes and making them produce nonfunctional proteins. Like I said, it is always hard to live with somebody. But some archaeon hosts must have managed to come to some kind of living arrangement with their new mitochondrial roommates that allowed the both of them to live together in a love-hate relationship that worked.
Modern eukaryotes have a distinctive nuclear membrane surrounding their DNA and the spliceosomes that splice out introns from mRNA work inside of this nuclear membrane to splice out the nasty introns within the mRNA. So initially, when a gene composed of DNA with embedded introns is transcribed to mRNA, everything is transcribed, including the introns, producing a strand of mRNA with "good" exon segments and "bad" intron segments. This happens all within the protection of the nuclear membrane that keeps the sexually turned on ribosomes, that are just dying to translate mRNA into proteins, at bay, like an elderly prom chaperone keeping things from getting out of hand. The spliceosomes then go to work within the nuclear membrane to splice out the parasitic introns, forming a new strand of mRNA that just contains the "good" exons. This highly edited strand of DNA then passes through a pore in the nuclear membrane out to the ribosomes patiently waiting outside - see Figure 12.
The nuclear membrane of modern eukaryotes provides the clue as to what happened. When the initial bacterial mitochondrial parasites would die inside of an archaeon host, they would simply dump all of their DNA into the interior of the archaeon host, and this allowed the parasitic bacterial introns in the dumped DNA to easily splice themselves at random points into the archaeon host DNA loop, and they frequently did so right in the middle of an archaeon host gene. Most times, that simply killed the archaeon host and all of its parasitic mitochondrial bacteria. Now the easiest way to prevent that from happening would be to simply put a membrane barrier around the archaeon host DNA loop to protect if from all of the dumped mitochondrial introns, and here is an explanation of how that could happen. It turns out that archaea and bacteria use different lipids to form their membranes, and although we know that the eukaryotes split off from the archaea, the eukaryotes strangely use bacterial lipids in their membranes instead of archaeon lipids. So the eukaryotes had to have transitioned from archaeon lipids to bacterial lipids at some point in time. The theory is that the genes for building bacterial lipids were originally on the bacterial mitochondrial invaders, but were later transplanted to the host archaeon DNA loops at some point. Once on the host archaeon DNA loop, those genes would then start to create bacterial lipids with no place to go. Instead, the generated lipids would simply form lipid "bags" near the host archaeon DNA loop. Those bacterial lipid "bags" would then tend to flatten, like empty plastic grocery bags, and then surround the host archaeon DNA loop. These flattened "bags" of bacterial lipids then evolved to produce the distinctive double-membrane structure of the nuclear membrane.
Figure 12 – The eukaryotic nuclear membrane is a double-membrane consisting of an inner and outer membrane separated by a perinuclear space. The nuclear membrane contains nuclear pores that allow edited mRNA to pass out but prevents ribosomes from entering and translating unedited mRNA containing introns into nonfunctional proteins. This double membrane is just the remnant of flattened "bags" of bacterial lipids that shielded the central archaeon DNA loop from the onslaught of parasitic mitochondrial DNA introns.
Nick Lane then goes on to explain how the host archaeon prokaryotic cells, trying to live with their parasitic mitochondrial bacterial roommates, and their unwelcome bacterial introns also led to the exclusive development of two sexes for eukaryotic-based life, and the use of sexual reproduction between those two sexes as well. It seems that the overwhelming benefit of having mitochondrial parasites, generating huge amounts of energy for the host archaeon prokaryotic cells, was just too much to resist, and the host archaeon prokaryotic cells went on to extremes to accommodate them, like also taking the host archaeon DNA loop that was initially wrapped around histone proteins to protect the DNA from the harsh environments that the extremophiles loved, and transforming that DNA into chromatin and chromosomes - for more on that see An IT Perspective on the Origin of Chromatin, Chromosomes and Cancer.
Nick Lane's key point is that the overwhelming amount of free energy that the parasitic mitochondrial bacteria brought to their original prokaryotic archaeon hosts was well worth the effort of learning to cope with a chronic infection that has lasted for 2 billion years. Yes, in some sense the mitochondrial parasites were indeed a collection of pain-in-the-butt pests, but putting up with them was well worth the effort because they provided their hosts with 5,000 times as much free energy as they originally had. That much energy allowed their hosts to put up with a lot of grief, and in the process of learning to cope with these parasites, the archaeon hosts went on to build all of the complex structures of the eukaryotic cell. This is a compelling argument, but it is very difficult to reconstruct a series of events that happened almost 2 billion years ago. Perhaps some softwarephysics and the evolution of software over the past 76 years, or 2.4 billion seconds, could be of assistance. During that long period of software evolution, did software also have to overcome a similar extremely inhibitive limiting factor that at first prevented the rise of complex software? Well, yes it did.
Using the Evolution of Software as a Guide
On the Earth we have seen life go through three major architectural advances:
1. The origin of life about 4 billion years ago, probably in the alkaline hydrothermal vents of the early Earth, producing the prokaryotic cell architecture.
2. The rise of the complex eukaryotic cell architecture about 2 billion years ago.
3. The rise of multicellular organisms consisting of millions, or billions, of eukaryotic cells all working together in the Ediacaran about 635 million years ago.
As we have seen, the most difficult thing to explain in the long history of life on this planet is not so much the origin of life itself, but the origin of the very complex architecture of the eukaryotic cell. This is where the evolutionary history of software on this planet can be of some help.
The evolutionary history of software on the Earth has converged upon a very similar historical path through Design Space because software also had to battle with the second law of thermodynamics in a highly nonlinear Universe - see The Fundamental Problem of Software for more on that. Software progressed through these similar architectures:
1. The origin of simple unstructured prokaryotic software on Konrad Zuse's Z3 computer in May of 1941 - 2.4 billion seconds ago.
2. The rise of structured eukaryotic software in 1972 - 1.4 billion seconds ago.
3. The rise of object-oriented software (software using multicellular organization) in 1995 - 694 million seconds ago
For more details on the above evolutionary history of software see the SoftwarePaleontology section of SoftwareBiology. From the above series of events, we can easily see that there was indeed a very long period of time, spanning at least one billion seconds, between 1941 and 1972 when only simple unstructured prokaryotic software was to be found on the Earth. Then early in the 1970s, highly structured eukaryotic software appeared and became the dominant form of software. Even today, the highly structured eukaryotic architecture of the early 1970s can still be seen in the modern object-oriented architecture of software.
Now suppose some alien AI software were to land on this planet today, and being just as self-absorbed as we are with finding out the nature of origins, they found a planet that was nearly totally dominated by software, but with some carbon-based life forms still hanging around that had not yet gotten the word, and with no written history of how the earthly software came to be. How could they piece together the evolutionary history of this newfound galactic software? The first thing they would notice is that nearly all of the software used multicellular organization based upon object-oriented programming languages. Some digging through old documents would reveal that object-oriented programming had actually been around since 1962, but that it did not at first catch on. In the late 1980s, the use of the very first significant object-oriented programming language, known as C++, started to appear in corporate IT, but object-oriented programming really did not become significant in IT until 1995 when both Java and the Internet Revolution arrived at the same time. The key idea in object-oriented programming is naturally the concept of an object. An object is simply a cell. Object-oriented languages use the concept of a Class, which is a set of instructions for building an object (cell) of a particular cell type in the memory of a computer. Depending upon whom you cite, there are several hundred different cell types in the human body, but in IT we generally use many thousands of cell types or Classes in commercial software. For a brief overview of these concepts go to the webpage below and follow the links by clicking on them.
Lesson: Object-Oriented Programming Concepts
http://docs.oracle.com/javase/tutorial/java/concepts/index.html
A Class defines the data that an object stores in memory and also the methods that operate upon the object data. Remember, an object is simply a cell. Methods are like biochemical pathways that consist of many steps or lines of code. A public method is a biochemical pathway that can be invoked by sending a message to a particular object, like using a ligand molecule secreted from one object to bind to the membrane receptors on another object. This binding of a ligand to a public method of an object can then trigger a cascade of private internal methods within an object or cell.
Figure 13 – A Class contains the instructions for building an object in the memory of a computer and basically defines the cell type of an object. The Class defines the data that an object stores in memory and also the methods that can operate upon the object data.
Figure 14 – Above is an example of a Bicycle object. The Bicycle object has three private data elements - speed in mph, cadence in rpm, and a gear number. These data elements define the state of a Bicycle object. The Bicycle object also has three public methods – changeGears, applyBrakes, and changeCadence that can be used to change the values of the Bicycle object’s internal data elements. Notice that the code in the object methods is highly structured and uses code indentation to clarify the logic.
Figure 15 – Above is some very simple Java code for a Bicycle Class. Real Class files have many data elements and methods and are usually hundreds of lines of code in length.
Figure 16 – Many different objects can be created from a single Class just as many cells can be created from a single cell type. The above List objects are created by instantiating the List Class three times and each List object contains a unique list of numbers. The individual List objects have public methods to insert or remove numbers from the objects and also a private internal sort method that could be called whenever the public insert or remove methods are called. The private internal sort method automatically sorts the numbers in the List object whenever a number is added or removed from the object.
Figure 17 – Objects communicate with each other by sending messages. Really one object calls the exposed public methods of another object and passes some data to the object it calls, like one cell secreting a ligand molecule that then plugs into a membrane receptor on another cell.
Figure 18 – In a growing embryo, the cells communicate with each other by sending out ligand molecules called morphogens, or paracrine factors, that bind to the membrane receptors on other cells.
Figure 19 – Calling a public method of an object can initiate the execution of a cascade of private internal methods within the object. Similarly, when a paracrine factor molecule plugs into a receptor on the surface of a cell, it can initiate a cascade of internal biochemical pathways. In the above figure, an Ag protein plugs into a BCR receptor and initiates a cascade of biochemical pathways or methods within a cell.
When a high-volume corporate website, consisting of many millions of lines of code running on hundreds of servers, starts up and begins taking traffic, billions of objects (cells) begin to be instantiated in the memory of the servers in a manner of minutes and then begin to exchange messages with each other in order to perform the functions of the website. Essentially, when the website boots up, it quickly grows to a mature adult through a period of very rapid embryonic growth and differentiation, as billions of objects are created and differentiated to form the tissues of the website organism. These objects then begin exchanging messages with each other by calling public methods on other objects to invoke cascades of private internal methods which are then executed within the called objects - for more on that see Software Embryogenesis.
In addition to finding lots of object-oriented software using millions or billions of intercommunicating objects, the aliens would also find some very old living fossils of huge single-celled Cobol programs still running on mainframe computers. The first Cobol program ran on December 6, 1960, on an RCA 501 computer, the first computer to use all transistors and no vacuum tubes. The RCA 501 came with 16K (131,072 bits) - 260K (2,129,920 bits) of magnetic core memory. The United States Air Force purchased an RCA 501 in 1959 for $121,698, which would now be $1,022,614 in 2017 dollars. I just checked, and you can now buy a laptop online with 4 GB of memory for $170. That 4 GB comes to 34,360,000,000 bits of computer memory, which is a little over 16,000 times as much memory as a fully loaded RCA 501 maxed out with 260K of memory. That comes to a total computer memory price-performance improvement of just a little over 97 million since 1959.
The Cobol programs written in the 1960s during the Unstructured Period used simple unstructured prokaryotic code because of the severe computer memory limitations of the computers of the day, but later in the 1970s and 1980s Cobol programs grew to become a single HUGE object, or cell, like a single-celled Paramecium, that allocated tons of memory up front and then processed the data read into the allocated memory with methods that are called subroutines.
Figure 20 – Cobol programs originally consisted of a few hundred lines of unstructured prokaryotic code, due to the severe memory limitations of the computers of the day, but in the 1970s and 1980s they grew to HUGE sizes of tens of thousands of lines of code using structured eukaryotic code, like a HUGE single-celled Paramecium, because cheap semiconductor memory chips dramatically increased the amount of memory computers could access.
Below is a code snippet from a fossil Cobol program listed in a book published in 1975. Notice the structured programming use of indented code and calls to subroutines with PERFORM statements.
PROCEDURE DIVISION.
OPEN INPUT FILE-1, FILE-2
PERFORM READ-FILE-1-RTN.
PERFORM READ-FILE-2-RTN.
PERFORM MATCH-CHECK UNTIL ACCT-NO OF REC-1 = HIGH_VALUES.
CLOSE FILE-1, FILE-2.
MATCH-CHECK.
IF ACCT-NO OF REC-1 < ACCT-NO OF REC-2
PERFORM READ-FILE-1-RTN
ELSE
IF ACCT-NO OF REC-1 > ACCT-NO OF REC-2
DISPLAY REC-2, 'NO MATCHING ACCT-NO'
PERORM READ-FILE-1-RTN
ELSE
PERORM READ-FILE-2-RTN UNTIL ACCT-NO OF REC-1
NOT EQUAL TO ACCT-NO OF REC-2
Figure 21 – A fossil Cobol program listed in a book published in 1975. Notice the structured programming use of indented code and calls to subroutines with PERFORM statements.
The aliens would also find a small amount of unstructured software, primarily in Unix shell scripts, but even that software would be much more structured than the code from before 1972. With a little more digging, the aliens would also find some truly ancient fossilized code. For example, below is a code snippet from a fossil Fortran program, listed in a book published in 1969, showing what ancient unstructured prokaryotic software really looked like. It has no internal structure and notice the use of GOTO statements to skip around in the code. Later this would become known as the infamous “spaghetti code” of the Unstructured Period that was such a joy to support.
30 DO 50 I=1,NPTS
31 IF (MODE) 32, 37, 39
32 IF (Y(I)) 35, 37, 33
33 WEIGHT(I) = 1. / Y(I)
GO TO 41
35 WEIGHT(I) = 1. / (-1*Y(I))
37 WEIGHT(I) = 1.
GO TO 41
39 WEIGHT(I) = 1. / SIGMA(I)**2
41 SUM = SUM + WEIGHT(I)
YMEAN = WEIGHT(I) * FCTN(X, I, J, M)
DO 44 J = 1, NTERMS
44 XMEAN(J) = XMEAN(J) + WEIGHT(I) * FCTN(X, I, J, M)
50 CONTINUE
Figure 22 – A fossil Fortran program, listed in a book published in 1969, showing what ancient unstructured prokaryotic software looked like. It has no internal structure and notice the use of GOTO statements to skip around in the code. Later this would become known as the infamous “spaghetti code” of the Unstructured Period that was such a joy to support.
The Extremely Inhibitive Limiting Factor of Software Complexity
Nick Lane argues that the extremely inhibitive limiting factor of biological complexity, that kept living things stuck at the level of simple prokaryotic cells for 2 billion years, was the amount of available free energy. Once the mitochondrial invaders came along and boosted the amount of available free energy available to a cell by a factor of 5,000, the very large and very complicated eukaryotic cell architecture became possible. But what was the extremely inhibitive limiting factor of software complexity that limited software to a very simple unstructured prokaryotic architecture for a span of more than one billion seconds from 1941 to 1972? Well in IT we all know that the two limiting factors for software are always processing speed and computer memory, and I would argue that the extremely inhibitive limiting factor of software complexity was simply the amount of available free computer memory.
Software is currently still being exclusively written by human beings, but that will all change sometime in the next 50 years when AI software will also begin to write software, and software will finally be able to self-replicate all on its own. In the meantime, let’s get back to the original rise of complex software. All computers have a CPU that can execute a fundamental set of primitive operations that are called its instruction set. The computer’s instruction set is formed by stringing together a large number of logic gates that are now composed of transistor switches. For example, all computers have a dozen or so registers that are like little storage bins for temporarily storing data that is being operated upon. A typical primitive operation might be taking the binary number stored in one register, adding it to the binary number in another register, and putting the final result into a third register. Since computers can only perform operations within their instruction set, computer programs written in high-level languages like C, C++, Fortran, Cobol or Visual Basic that can be read by a human programmer, must first be compiled, or translated, into a file that consists of the “1s” and “0s” that define the operations to be performed by the computer in terms of its instruction set. This compilation, or translation process, is accomplished by feeding another compiled program, called a compiler, with the source code of the program to be translated. The output of the compiler program is called the compiled version of the program and is an executable file on disk that can be directly loaded into the memory of a computer and run. Computers also now have memory chips that can store these compiled programs and the data that the compiled programs process. For example, when you run a compiled program, by double-clicking on its icon on your desktop, it is read from disk into the memory of your computer, and it then begins executing the primitive operations of the computer’s instruction set as defined by the compiled program.
Figure 23 – A ribosome behaves very much like a compiler. It reads the mRNA source code and translates it into an executable polypeptide chain of amino acids that then go on to fold up into a 3-D protein molecule that can directly execute a biochemical function, much like a compiled executable file that can be directly loaded into the memory of a computer to do things.
Below is the source code for a simple structured program, with a single Average function, that computes the average of several numbers that are entered via the command line of a computer. Please note that modern applications now consist of many thousands to many millions of lines of structured code. The simple example below is just for the benefit of our non-IT readers to give them a sense of what is being discussed when I describe the compilation of source code into executable files that can be loaded into the memory of a computer and run.
Figure 24 – Source code for a C program that calculates an average of several numbers entered at the keyboard.
The problem is that the amount of free computer memory determines the maximum size of a program because the executable file for the program has to fit into the available free memory of the computer at runtime. Generally, the more lines of source code you have in your program, the larger will be its compiled executable file, so the amount of available free memory determines the maximum size of your program. Now in the 1970s people did come up with computer operating systems that could use virtual memory to remove this memory constraint, but that happened after structured eukaryotic software had appeared, so we can overlook that bit of IT history. Now in the 1950s and 1960s during the prokaryotic Unstructured Period, computer memory was very expensive and very limited. Prior to 1955 computers, like the UNIVAC I that first appeared in 1951, were using mercury delay lines that consisted of a tube of mercury that was about 3 inches long. Each mercury delay line could store about 18 bits of computer memory as sound waves that were continuously refreshed by quartz piezoelectric transducers on each end of the tube. Mercury delay lines were huge and very expensive per bit so computers like the UNIVAC I only had a memory of 12 K (98,304 bits).
Figure 25 – Prior to 1955, huge mercury delay lines built from tubes of mercury that were about 3 inches long were used to store bits of computer memory. A single mercury delay line could store about 18 bits of computer memory as a series of sound waves that were continuously refreshed by quartz piezoelectric transducers at each end of the tube.
In 1955 magnetic core memory came along, and used tiny magnetic rings called "cores" to store bits. Four little wires had to be threaded by hand through each little core in order to store a single bit, so although magnetic core memory was a lot cheaper and smaller than mercury delay lines, it was still very expensive and took up lots of space.
Figure 26 – Magnetic core memory arrived in 1955 and used a little ring of magnetic material, known as a core, to store a bit. Each little core had to be threaded by hand with 4 wires to store a single bit.
Figure 27 – Magnetic core memory was a big improvement over mercury delay lines, but it was still hugely expensive and took up a great deal of space within a computer.
Because of the limited amount of free computer memory during the 1950s and 1960s, computers simply did not have enough free computer memory to allow people to write very large programs, so programs were usually just a few hundred lines of code each. Now you really cannot do much logic in a few hundred lines of code, so IT people would string together several small programs into a batch run. Input-Output tapes were used between each small program in the batch run. The first small program would run and write its results to one or more output tapes. The next program in the batch run would then read those tapes and do some more processing, and write its results to one or more output tapes too. This continued on until the very last program in the batch run wrote out its final output tapes. For more on that see: An IT Perspective on the Origin of Chromatin, Chromosomes and Cancer. Now when you are writing very small programs, because you are severely limited by the available amount of free memory, there really is no need to write structured code because you are dealing with a very small amount of processing logic. So during the Unstructured Period of the 1950s and 1960s, IT professionals simply did not bother with breaking up software logic into functions or subroutines. So each little program in a batch stream was like a small single prokaryotic cell, with no internal structure.
But in the early 1970s, inexpensive semiconductor memory chips came along that made computer memory small and cheap. These memory chips were the equivalent of software mitochondria because they removed the extremely inhibitive limiting factor of software complexity. Suddenly, IT now had large amounts of computer memory that allowed IT people to write huge programs. But that presented a problem. It was found that the processing logic became too contorted and impossible to maintain when simple prokaryotic unstructured programs were scaled up to programs with 50,000 lines of unstructured code. Even the programmer who wrote the original code could not make sense of it a few months later, and this was much worse for new programmers who came along later to maintain the code. Since commercial software can easily live for 10 years or more, that was a real problem. To alleviate this problem, Dahl, Dijkstra, and Hoare published Structured Programming in 1972, in which they suggested that computer programs should have a complex internal structure with no GOTO statements, lots of subroutines, indented code, and many comment statements. During the Structured Period that soon followed, these structured programming techniques were adopted by the IT community, and the GOTO statements were replaced by subroutines, also known as functions(), and indented code with lots of internal structure, like the eukaryotic structure of modern cells that appeared about 2 billion years ago.
Figure 28 – Finally in the early 1970s inexpensive semiconductor memory chips came along that made computer memory small and cheap. These memory chips were the equivalent of software mitochondria because they removed the extremely inhibitive limiting factor of software complexity.
Today, we now almost exclusively use object-oriented programming techniques that allow for the multicellular organization of software via objects, but the code for the methods that operate on those objects, still use the structured programming techniques that first appeared in the early 1970s.
Conclusion
Now many tend to dismiss the biological findings of softwarephysics because software currently is a product of the human mind, while biological life is not a product of intelligent design. Granted, biological life is not a product of intelligent design, but neither is the human mind. The human mind and biological life are both the result of natural processes at work over very long periods of time. This objection simply stems from the fact that we are all still, for the most part, self-deluded Cartesian dualists at heart, with seemingly a little “Me” running around within our heads that just happens to have the ability to write software and to do other challenging things. Thus, most human beings do not think of themselves as part of the natural world. Instead, they think of themselves, and others, as immaterial spirits temporarily haunting a body, and when that body dies the immaterial spirit lives on. In this view, human beings are not part of the natural world. Instead, they are part of the supernatural. But in softwarephysics, we maintain that the human mind is a product of natural processes in action, and so is the software that it produces. For more on that see The Ghost in the Machine the Grand Illusion of Consciousness.
Still, I realize that there might be some hesitation to pursue this line of thought because it might be construed by some as an advocacy of intelligent design, but that is hardly the case. The evolution of software over the past 76 years has essentially been a matter of Darwinian inheritance, innovation and natural selection converging upon similar solutions to that of biological life. For example, it took the IT community about 60 years of trial and error to finally stumble upon an architecture similar to that of complex multicellular life that we call SOA – Service Oriented Architecture. The IT community could have easily discovered SOA back in the 1960s if it had adopted a biological approach to software and intelligently designed software architecture to match that of the biosphere. Instead, the worldwide IT architecture we see today essentially evolved on its own because nobody really sat back and designed this very complex worldwide software architecture; it just sort of evolved on its own through small incremental changes brought on by many millions of independently acting programmers through a process of trial and error. When programmers write code, they always take some old existing code first and then modify it slightly by making a few changes. Then they add a few additional new lines of code and test the modified code to see how far they have come. Usually, the code does not work on the first attempt because of the second law of thermodynamics, so they then try to fix the code and try again. This happens over and over until the programmer finally has a good snippet of new code. Thus, new code comes into existence through the Darwinian mechanisms of inheritance coupled with innovation and natural selection - for more on that see How Software Evolves. Some might object that this coding process of software is actually a form of intelligent design, but that is not the case. It is important to differentiate between intelligent selection and intelligent design. In softwarephysics we extend the concept of natural selection to include all selection processes that are not supernatural in nature, so for me, intelligent selection is just another form of natural selection. This is really nothing new. Predators and prey constantly make “intelligent” decisions about what to pursue and what to evade, even if those “intelligent” decisions are only made with the benefit of a few interconnected neurons or molecules. So in this view, the selection decisions that a programmer makes after each iteration of working on some new code really are a form of natural selection. After all, programmers are just DNA survival machines with minds infected with memes for writing software, and the selection processes that the human mind undergo while writing software are just as natural as the Sun drying out worms on a sidewalk or a cheetah deciding upon which gazelle in a herd to pursue.
For example, when IT professionals slowly evolved our current $10 trillion worldwide IT architecture over the past 2.4 billion seconds, they certainly did not do so with the teleological intent of creating a simulation of the evolution of the biosphere. Instead, like most organisms in the biosphere, these IT professionals were simply trying to survive just one more day in the frantic world of corporate IT. It is hard to convey the daily mayhem and turmoil of corporate IT to outsiders. When I first hit the floor of Amoco’s IT department back in 1979, I was in total shock, but I quickly realized that all IT jobs essentially boiled down to simply pushing buttons. All you had to do was to push the right buttons, in the right sequence, at the right time, and with zero errors. How hard could that be? Well, it turned out to be very difficult indeed, and in response, I began to subconsciously work on softwarephysics to try to figure out why this job was so hard, and how I could dig myself out of the mess that I had gotten myself into. After a while, it dawned on me that the fundamental problem was the second law of thermodynamics operating in a nonlinear simulated universe. The second law made it very difficult to push the right buttons in the right sequence and at the right time because there were so many erroneous combinations of button pushes. Writing and maintaining software was like looking for a needle in a huge utility phase space. There just were nearly an infinite number of ways of pushing the buttons “wrong”. The other problem was that we were working in a very nonlinear utility phase space, meaning that pushing just one button incorrectly usually brought everything crashing down. Next, I slowly began to think of pushing the correct buttons in the correct sequence as stringing together the correct atoms into the correct sequence to make molecules in chemical reactions that could do things. I also knew that living things were really great at doing that. Living things apparently overcame the second law of thermodynamics by dumping entropy into heat as they built low entropy complex molecules from high entropy simple molecules and atoms. I then began to think of each line of code that I wrote as a step in a biochemical pathway. The variables were like organic molecules composed of characters or “atoms” and the operators were like chemical reactions between the molecules in the line of code. The logic in several lines of code was the same thing as the logic found in several steps of a biochemical pathway, and a complete function was the equivalent of a full-fledged biochemical pathway in itself. For more on that see Some Thoughts on the Origin of Softwarephysics and Its Application Beyond IT and SoftwareChemistry.
So based on the above analysis, I have a high level of confidence that Nick Lane has truly solved the greatest mystery in biology, and that is indeed quite an accomplishment!
Comments are welcome at scj333@sbcglobal.net
To see all posts on softwarephysics in reverse order go to:
https://softwarephysics.blogspot.com/
Regards,
Steve Johnston
























