One of my most favorite scientific YouTubers is Anton Petrov. Anton reads tons of scientific papers and somehow manages to produce a new YouTube video every day covering some of his latest reads. His videos come out so frequently, that it is hard to keep up, but they are always very excellent. I recently watched:
Molecule of Life Found In a Cloud Near Center of Milky Way
https://www.youtube.com/watch?v=TCl0TtoJDD0&t=1s
which covers this paper:
Discovery in space of ethanolamine, the simplest phospholipid head group
https://www.pnas.org/content/118/22/e2101314118
Now, why should IT professionals be interested in phospholipid head groups? This is an important discovery because phospholipid head groups make phospholipid bilayers possible and they form the structural foundations of all carbon-based life on the Earth. This is also an important discovery for the origin and early evolution of computer software because software first arose in the Minds of carbon-based human DNA survival machines. And if we finally ever do find some alien software out there, it too will probably have arisen from carbon-based life on some distant world. So all IT professionals should naturally be very interested in the origins of carbon-based life. In my last posting The Evolution of Software As Seen Through the Lens of Geological Deep Time, we saw that carbon-based life can be thought of as the software of the Earth's crust and that the rocks of the Earth's crust could also be thought of as the hardware upon which that carbon-based software runs. We also saw that the evolution of computer software followed a very similar path through Design Space over the past 80 years, or 2.5 billion seconds, as did carbon-based life on the Earth over the past 4.0 billion years. This resulted from both IT professionals and carbon-based life converging on very similar solutions to common data processing problems guided by the Darwinian mechanisms of inheritance, innovation and natural selection. Galactic molecular clouds are important in all of this because all of the atoms and molecules of the Earth's crust originally came from the galactic molecular cloud that formed the solar system and several thousand other nearby stars in the original open cluster of stars that our Sun formed in. Some of those atoms and molecules came early, and some of them came later. The late arrivers were ferried here onboard impacting asteroids and comets. Still, all the atoms and molecules of the Earth's crust, and all of the atoms in your computer, originally came from the galactic molecular cloud that formed the solar system.
In The Evolution of Software As Seen Through the Lens of Geological Deep Time, we also saw how computer software evolved over the past 2.5 billion seconds by exapting the switching technology from telephone relays, radio and TV vacuum tubes, portable radio transistors and integrated circuit chips from the consumer electronics industry. Similarly, in order for carbon-based life to get a start, it had to exapt the existing atoms and molecules that it had at hand on the surface of the Earth. So it is important to know exactly how those molecules formed and where they came from, and that is the topic of the above paper. Exapting technology used for one purpose to then perform a new purpose is certainly a hallmark of all forms of self-replicating information like carbon-based life and computer software.
Once again, let me repeat the fundamental characteristics of self-replicating information for those of you new to softwarephysics.
Self-Replicating Information – Information that persists through time by making copies of itself or by enlisting the support of other things to ensure that copies of itself are made.
The Characteristics of Self-Replicating Information
All forms of self-replicating information have some common characteristics:
1. All self-replicating information evolves over time through the Darwinian processes of inheritance, innovation and natural selection, which endows self-replicating information with one telling characteristic – the ability to survive in a Universe dominated by the second law of thermodynamics and nonlinearity.
2. All self-replicating information begins spontaneously as a parasitic mutation that obtains energy, information and sometimes matter from a host.
3. With time, the parasitic self-replicating information takes on a symbiotic relationship with its host.
4. Eventually, the self-replicating information becomes one with its host through the symbiotic integration of the host and the self-replicating information.
5. Ultimately, the self-replicating information replaces its host as the dominant form of self-replicating information.
6. Most hosts are also forms of self-replicating information.
7. All self-replicating information has to be a little bit nasty in order to survive.
8. The defining characteristic of self-replicating information is the ability of self-replicating information to change the boundary conditions of its utility phase space in new and unpredictable ways by means of exapting current functions into new uses that change the size and shape of its particular utility phase space. See Enablement - the Definitive Characteristic of Living Things for more on this last characteristic. That posting discusses Stuart Kauffman's theory of Enablement in which living things are seen to exapt existing functions into new and unpredictable functions by discovering the “AdjacentPossible” of springloaded preadaptations.
Over the past 4.56 billion years we have seen five waves of self-replicating information sweep across the surface of the Earth and totally rework the planet, as each new wave came to dominate the Earth:
1. Self-replicating autocatalytic metabolic pathways of organic molecules
2. RNA
3. DNA
4. Memes
5. Software
Software is currently the most recent wave of self-replicating information to arrive upon the scene and is rapidly becoming the dominant form of self-replicating information on the planet. For more on the above see A Brief History of Self-Replicating Information. But to fully understand the origin of software in the Universe, we need to first understand the origin of carbon-based life.
The Origin of Carbon-Based Life on the Earth
In The Bootstrapping Algorithm of Carbon-Based Life, we covered the Hot Spring Origins Hypothesis of Dave Deamer and Bruce Damer out of the University of California at Santa Cruz. The Hot Spring Origins Hypothesis proposes that carbon-based life did not originate in the sea but originated on dry land in hydrothermal pools.
Figure 1 – Above is Bumpass Hell, a hydrothermal field on the volcanic Mount Lassen in California that Dave Deamer and Bruce Damer cite as a present-day example of the type of environment that could have brought forth carbon-based life about four billion years ago.
.
Figure 2 – Dave Deamer's and Bruce Damer's new Hot Spring Origins Hypothesis proposes that when a bathtub ring around a hydrothermal pool periodically dries out, the resulting desiccation chemically squeezes out water molecules between monomers, causing them to be glued together into polymers. Then the hydrothermal pool refills with water and the process repeats. This may be the naturally occurring bootstrapping algorithm for carbon-based life throughout our Universe.
Bruce Damer was kind enough to send me a PDF of their poster from the 2016 ISSOL - The International Society for the Study of the Origin of Life - meeting that features all of the stages of their Hot Spring Origins Hypothesis. Please note that the color-coded text corresponds to the color-coded sections on the poster image.
A Hot Spring Origin of Life and Early Adaption Pathway in seven steps:
1. Synthesis of organics (a), key primordial building blocks for life, occurs in space prior and during the formation of the Solar System;
2. Accumulation of in-falling organics and other compounds generated within hot springs on an active volcanic landscape combine and undergo self-assembly of structures such as lipid membranes;
3. Concentration of compounds in small pools utilizes sunlight, heat and chemical energy to drive key prebiotic polymerization reactions and self-assembly of membranous structures;
4. Cycling of the products of these reactions in a wet-dry fluctuating hot spring ‘origin pool’ drives them through three phases: organic membranes in the pool i) Dry down to form layered Films between which organic building blocks bond together to form polymers; on refilling the films ii) Wet and bud off trillions of lipid Protocells some encapsulating random polymers. Each protocell undergoes a iii) Test, the first form of natural selection, and stable survivors accumulate into a moist Gel as the pool level drops. Through many iterations polymers iv) Interact, within the Gel and the Films evolving ever more complex functions until a form of pre-life (b) called a Progenote emerges that is able to grow and adapt;
5. Distribution of robust progenotes occurs by water or wind to other pools, rivers, and lakes, where they acquire and share evolutionary innovations including an early form of photosynthesis. Eventually, protocells develop the complicated innovation of cell division and initiate the transition into early life (c);
6. Adaptation of early microbial communities to stressful saltwater estuaries prepares them for access to the more extreme marine environment;
7. Large tides at the ancient seacoast select for microbial communities able to cement sand grains together forming the layers that build the stromatolites so abundant in the fossil record. Global life (d) enables Colonization of many niches on the land and in the sea, setting the stage for free-living cells and, after billions of years, complex multi-cellular organisms.
Dave Deamer has been working on the origin of carbon-based life for more than 45 years. He is definitely of the "cell membranes came first" school of thought that argues that carbon-based life first required some kind of container to get started. This original container separated the inside of proto-cells from the outside of proto-cells and provided a mechanism to isolate the functions of carbon-based life from the environment in which early proto-cells found themselves.
For example, all current carbon-based cells have at least one large membrane that surrounds the entire cell. The cell membrane consists of a phospholipid bilayer consisting of molecules with a water-loving hydrophilic phospholipid head and a set of water-hating hydrophobic phospholipid tails as shown in Figure 3. Embedded in this phospholipid bilayer are other molecules that allow the inside of a cell to communicate with the outside of the cell in a controlled manner. This is why finding preformed phospholipid head molecules in galactic molecular clouds is important. The Hot Spring Origins Hypothesis proposes that the primordial molecules of life were formed in interstellar space prior to the origination of the Earth with no need to form them from the primitive atoms of the Earth. Below is a list of such molecules.
List of interstellar and circumstellar molecules
https://en.wikipedia.org/wiki/List_of_interstellar_and_circumstellar_molecules
Figure 3 – A cell membrane consists of a phospholipid bilayer with embeded molecules that allow for a controlled input-output to the cell.
But how do you create these highly ordered phospholipid bilayer membranes from individual phospholipid molecules? At first glance, a highly ordered phospholipid bilayer looks like something that would be hard to make and maintain because its highly ordered appearance would seem to make it a very low entropy construction. The second law of thermodynamics should demand that some energy be expended to build and maintain such a wall. And it does, but not the way you might think. Water molecules are polar molecules that have a positive end and a negative end because oxygen atoms attract the bonding electrons more strongly than do the hydrogen atoms. The positive ends of water molecules attract the negative ends of other water molecules to form a loosely coupled network of water molecules with a minimum of free energy. This loosely coupled network of water molecules would also seem to have a lower entropy than a truly random configuration of water molecules and also a violation of the second law of thermodynamics. But in reality, some of the free energy of the water molecules was turned into the random motions of heat energy when the water molecules rotated to form the network producing a total increase of entropy in the Universe as the second law of thermodynamics demands. Since phospholipid bilayers are surrounded by polar water molecules on both sides, the polar phospholipid heads are similarly drawn to the polar water molecules to minimize their free energies. Thus, phospholipid bilayers naturally form when phospholipid molecules are added to water. For more on the second law of thermodynamics and information and why the second law makes coding software so difficult see Entropy - the Bane of Programmers and The Demon of Software.

Figure 4 – Water molecules are polar molecules that have a positive end and a negative end because oxygen atoms attract the binding electrons more strongly than do the hydrogen atoms. The positive ends of water molecules attract the negative ends of other water molecules to form a loosely coupled network of water molecules with a minimum of free energy.
You can easily demonstrate the making of phospholipid bilayers the next time you make some toast. After you butter the toast, you will find a greasy layer of butter all over your knife that will not wash off when you run the knife under the kitchen faucet. Now run some water over your fingers and then rub them on the bar of soap at your kitchen sink. Next, take your wet soapy fingers and rub them all over the greasy layer of butter on your knife. Now run the knife and your fingers under the kitchen faucet. Amazingly, your knife will now be totally clean with no residual grease at all when you dry it off! This is because the lipids in the bar of soap have surrounded a huge number of little drops of greasy butter molecules with a lipid membrane called a micelle that has its water-loving phospholipid heads on the outside. This creates a huge number of little lipid vesicles enclosing greasy butter molecules. That is how soap works. The lipids break the high water surface tension of the polar water molecules, and they create grease-cutting micelle vesicles too.
Figure 5 – The lipids in a bar of soap have water-loving polar heads and water-hating nonpolar tails. The soap lipids can form a spherical micelle with all of the water-hating nonpolar tails facing inwards. A spherical micelle can surround the greasy nonpolar molecules of butter and allow them to be flushed away by a stream of polar water molecules. The lipids in a bar of soap can also form a cell-like liposome with a bilayer of lipid molecules that can surround the monomers and polymers of life.
In the Hot Spring Origins Hypothesis phospholipid molecules from interstellar space become concentrated in hydrothermal pools due to the constant evaporation of hot water. The phospholipid molecules then naturally form cell-like liposomes with a bilayer of lipid molecules that can surround the monomers and polymers of life. Periodic desiccation of the hydrothermal pools chemically squeezes out water molecules between monomers to form new polymers. When the hydrothermal pools are refilled by rainwater, the process repeats. This process could repeat for millions of years until a self-replicating proto-cell formed.
Figure 6 – The prokaryotic cell architecture of the bacteria and archaea contains just one large membrane surrounding the entire cell. Eukaryotic cells, on the other hand, have many additional internal membranes to isolate the logical operations of the cell, such as membranes for the nuclear envelope, the endoplasmic reticulum, mitochondria, chloroplasts, Golgi bodies, lysosomes and vesicles.
As you can see, cell membranes have played a critical role in the development of carbon-based life. Now let's look for some similarities in the evolution of software.
The Importance of Functional Isolation by Membranes in the Evolution of Software
During the Unstructured Period (1941 – 1972) the functionality found within software was naturally isolated into small chunks of logic because of the very limited amounts of computer memory at the time. At the time, million-dollar mainframes might have a total of 1 MB of memory and that memory was divided up into several 128 KB and 256 KB chunks. So programs were limited to run in a chunk of memory that was only 128 KB to 256 KB in size and you cannot do a lot of logic in that small amount of memory. So the logic for an entire Application was divided into chunks that could fit into 128 KB to 256 KB of memory. Applications were run as batch jobs with a large number of steps. Each step ran a small program that could fit into the 128 KB to 256 KB of memory and input and output tapes were used between each job step to ferry along the data being processed. For more on batch tape processing see An IT Perspective on the Origin of Chromatin, Chromosomes and Cancer.
Figure 7 – During the Unstructured Period (1941 – 1972), software logic was naturally broken up into small chunks that could fit into 128 KB to 256 KB of memory. The steps in a job stream used lots of input and output tapes to ferry along the data being processed. Each little program in a job stream was surrounded by its own little isolating membrane like the early prokaryotic bacteria and archaea.
During the Structured Period (1972 – 1992), the amount of memory available on mainframes exploded with the introduction of memory chips. For more on that see The Rise of Complexity in Living Things and Software. People then began to write much larger unstructured programs consisting of huge amounts of "spaghetti code" that could not be maintained. In response, structured programming was invented to break up the unstructured "spaghetti code" logic into a large number of functions() or subroutines() that could be called from the mainline() program. Each function() or subroutine() was surrounded by its own little membrane that isolated the code in the function() or subroutine() from the code in the mainline() program. Data entering the functions() or subroutines() from the mainline() could be checked for errors before processing and could be also checked before returning to the mainline(). This isolation of logic by membranes made very large programs possible and greatly eased their maintenance.
During the Object-Oriented Period (1992 – Present), programmers adopted a multicellular organization for software, in which programs consisted of many instances of objects (cells) that were surrounded by membranes studded with exposed methods (membrane receptors). Some investigation reveals that object-oriented programming has actually been around since 1962, but that it did not catch on at first. In the late 1980s, the use of the very first significant object-oriented programming language, known as C++, started to appear in corporate IT, but object-oriented programming really did not become significant in IT until 1995 when both Java and the Internet Revolution arrived at the same time. The key idea in object-oriented programming is naturally the concept of an object. An object is simply a cell. Object-oriented languages use the concept of a Class, which is a set of instructions for building an object (cell) of a particular cell type in the memory of a computer. Depending upon whom you cite, there are several hundred different cell types in the human body, but in IT we generally use many thousands of cell types or Classes in commercial software. For a brief overview of these concepts go to the webpage below and follow the links by clicking on them.
Lesson: Object-Oriented Programming Concepts
http://docs.oracle.com/javase/tutorial/java/concepts/index.html
A Class defines the data that an object stores in memory and also the methods that operate upon the object data. Remember, an object is simply a cell. Methods are like biochemical pathways that consist of many steps or lines of code. A public method is a biochemical pathway that can be invoked by sending a message to a particular object, like using a ligand molecule secreted from one object to bind to the membrane receptors on another object. This binding of a ligand to a public method of an object can then trigger a cascade of private internal methods within an object or cell.
Figure 8 – A Class contains the instructions for building an object in the memory of a computer and basically defines the cell type of an object. The Class defines the data that an object stores in memory and also the methods that can operate upon the object data.
Figure 9 – Above is an example of a Bicycle object. The Bicycle object has three private data elements - speed in mph, cadence in rpm, and a gear number. These data elements define the state of a Bicycle object. The Bicycle object also has three public methods – changeGears, applyBrakes, and changeCadence that can be used to change the values of the Bicycle object’s internal data elements. Notice that the code in the object methods is highly structured and uses code indentation to clarify the logic.
Figure 10 – Above is some very simple Java code for a Bicycle Class. Real Class files have many data elements and methods and are usually hundreds of lines of code in length.
Figure 11 – Many different objects can be created from a single Class just as many cells can be created from a single cell type. The above List objects are created by instantiating the List Class three times and each List object contains a unique list of numbers. The individual List objects have public methods to insert or remove numbers from the objects and also a private internal sort method that could be called whenever the public insert or remove methods are called. The private internal sort method automatically sorts the numbers in the List object whenever a number is added or removed from the object.
Figure 12 – Objects communicate with each other by sending messages. Really one object calls the exposed public methods of another object and passes some data to the object it calls, like one cell secreting a ligand molecule that then plugs into a membrane receptor on another cell.
Figure 13 – In a growing embryo, the cells communicate with each other by sending out ligand molecules called morphogens, or paracrine factors, that bind to the membrane receptors on other cells.
Figure 14 – Calling a public method of an object can initiate the execution of a cascade of private internal methods within the object. Similarly, when a paracrine factor molecule plugs into a receptor on the surface of a cell, it can initiate a cascade of internal biochemical pathways. In the above figure, an Ag protein plugs into a BCR receptor and initiates a cascade of biochemical pathways or methods within a cell.
When a high-volume corporate website, consisting of many millions of lines of code running on hundreds of servers, starts up and begins taking traffic, billions of objects (cells) begin to be instantiated in the memory of the servers in a manner of minutes and then begin to exchange messages with each other in order to perform the functions of the website. Essentially, when the website boots up, it quickly grows to a mature adult through a period of very rapid embryonic growth and differentiation, as billions of objects are created and differentiated to form the tissues of the website organism. These objects then begin exchanging messages with each other by calling public methods on other objects to invoke cascades of private internal methods which are then executed within the called objects - for more on that see Software Embryogenesis.
Object-Oriented Class Libraries
In addition to implementing a form of multicellular organization in software, the object-oriented programming languages, like C++ and Java, brought with them the concept of a class library. A class library consists of the source code for a large number of reusable Classes for a multitude of objects. For example, for a complete list of the reusable objects in the Java programming language see:
Java Platform, Standard Edition 7
API Specification
https://docs.oracle.com/javase/7/docs/api/
The "All Classes" pane in the lower left of the webpage lists all of the built-in classes of the Java programming language.
Figure 15 – All of the built-in Classes of the Java programming language inherit the data elements and methods of the undifferentiated stem cell Object.
The reusable classes in a class library all inherit the characteristics of the fundamental class Object. An Object object is like a stem cell that can differentiate into a large number of other kinds of objects. Below is a link to the Object Class and all of its reusable methods:
Class Object
https://docs.oracle.com/javase/7/docs/api/java/lang/Object.html
For example, a String object is a particular object type that inherits all of the methods of a general Object stem cell and can perform many operations on the string of characters it contains via the methods of a String object.
Figure 16 – A String object contains a number of characters in a string. A String object inherits all of the general methods of the Object Class, and contains many additional methods, like finding a substring in the characters stored in the String object.
The specifics of a String object can be found at:
Class String
https://docs.oracle.com/javase/7/docs/api/java/lang/String.html
Take a look at the Method Summary of the String class. It contains about 50 reusable code methods that can perform operations on the string of characters in a String object. Back in the unstructured prokaryotic times of the 1950s and 1960s, the source code to perform those operations was repeatedly reprogrammed over and over because there was no easy way to reuse source code in those days. But in the multicellular object-oriented period, the use of extensive class libraries allowed programmers to easily assemble the "conserved core processes" that had been developed back in the unstructured prokaryotic source code of the 1950s and 1960s into complex software by simply instantiating objects and making calls to the methods of the objects that they needed. Thus, the new multicellular object-oriented source code, that created and extended the classes found within the already existing class library, took on more of a regulatory nature than the source code of old. This new multicellular object-oriented source code carried out its required logical operations by simply instantiating certain objects in the class library of reusable code, and then choosing which methods needed to be performed on those objects.
Finally, the SOA - Service Oriented Architecture Period (2004 – 2015) and the Cloud Computing Microservices Period (2015 - Present) of today further relied on object-oriented membranes to isolate functionality that could be shared across a network by allowing objects on one computer to call the methods() of objects on another network server or from a Cloud Container via the Internet.
Conclusion
Since the very beginning, the architecture of software has evolved through the Darwinian processes of inheritance, innovation and natural selection and has followed a path very similar to the path followed by the carbon-based living things on the Earth. I believe this has been due to what evolutionary biologists call convergence. For example, the concept of the eye has independently evolved at least 40 different times in the past 600 million years, and there are many examples of “living fossils” showing the evolutionary path. For example, the camera-like structures of the human eye and the eye of an octopus are nearly identical, even though each structure evolved totally independent of the other.
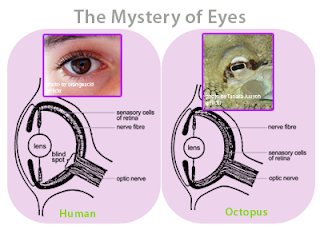
Figure 17 - The eye of a human and the eye of an octopus are nearly identical in structure, but evolved totally independently of each other. As Daniel Dennett pointed out, there are only a certain number of Good Tricks in Design Space and natural selection will drive different lines of descent towards them.
Figure 18 – There are many living fossils that have left behind signposts along the trail to the modern camera-like eye. Notice that the human-like eye on the far right is really that of an octopus (click to enlarge).
An excellent treatment of the significance that convergence has played in the evolutionary history of life on Earth, and possibly beyond, can be found in Life’s Solution (2003) by Simon Conway Morris. The convergent evolution for carbon-based life on the Earth to develop eyes was driven by the hardware fact of life that the Earth is awash in solar photons.
Programmers and living things both have to deal with the second law of thermodynamics and nonlinearity, and there are only a few optimal solutions. Programmers try new development techniques, and the successful techniques tend to survive and spread throughout the IT community, while the less successful techniques are slowly discarded. Over time, the population distribution of software techniques changes. The invention of the membrane by both carbon-based life and software to provide a mechanism to isolate functionality is just another example of such convergence in action.
Comments are welcome at scj333@sbcglobal.net
To see all posts on softwarephysics in reverse order go to:
https://softwarephysics.blogspot.com/
Regards,
Steve Johnston


















No comments:
Post a Comment