In Programming Biology in the Biological Computation Group of Microsoft Research we saw that Microsoft is currently working on a biological IDE (Integrated Development Environment) called Station B for biologists that would work much like Eclipse or Microsoft's Visual Studio does for computer software developers. The purpose of Station B is to heavily automate the very tedious process of understanding what old DNA source code does and editing it with new DNA in order to be able to program biology. In order to do that, Microsoft is leveraging its 30 years of experience with developing and maintaining software source code. Now, modern software developers might wonder what's the big deal with that? That is because modern software developers never suffered with the limitations of programming computer source code on punch cards. And sadly, until very recently, biologists have been essentially forced to do biological programming on cards. Only in the past few years have biological programmers developed the CRISPR technology that lets them edit DNA source code with something that is the equivalent of an early 1970s-like line editor like ed or ex on Unix.
So in solidarity with our new-found biological programmers, let's review the history of how software source code has been developed and maintained over the past 79 years, or 2.5 billion seconds, ever since Konrad Zuse first cranked up his Z3 computer in May of 1941. For more on the computational adventures of Konrad Zuse please see So You Want To Be A Computer Scientist?. Unfortunately, even with the new CRISPR technology, synthetic biology programmers are currently still essentially stuck back in the 1970s when it comes to editing DNA source code. The purpose of this posting is to let them know that computer software programmers have been there too and that there is the possibility for great improvements in the future. But first, let's see how CRISPR works from a computer programmer's perspective.
How CRISPR Works
In 2012, Jennifer Doudna and Emmanuelle Charpentier published a paper that described how DNA could be precisely edited using a CRISPR-Cas9 complex that could be programmed with specific strings of RNA to edit targeted DNA in cells. Similarly, in 2016 it was discovered that a CRISPR-Cas13 complex could edit targeted RNA. For the first time, CRISPR technology now allows biologists to find specific strings of DNA bases in cells and replace them with new strings of DNA bases. This allows biologists to knock out specific genes, modify specific genes or even add new genes to eukaryotic cells. CRISPR is essentially a line editor that allows biologists to now locate specific lines of code in DNA and make a change to that line of code. CRISPR was actually invented by bacteria as a literal form of antiviral software that operates exactly like the antivirus software we run on our hardware. A virus is a parasitic form of DNA or RNA wrapped in a protein coat that parasitizes cells including bacterial cells. For more about viruses see A Structured Code Review of the COVID-19 Virus. Viruses that infect bacteria are called phages. After a viral phage attaches itself to a bacterial cell, it injects its DNA or RNA into the bacterial cell. The DNA or RNA of the viral phage then takes over the machinery of the bacterial cell and has the bacterial cell create hundreds of copies of the viral phage that either leak out of the bacterial cell or cause the bacterial cell to burst and die. So a virus is a pure form of parasitic self-replicating information. For more about the vast powers of self-replicating information see A Brief History of Self-Replicating Information.
Now billions of years ago, bacteria came up with a defense mechanism against viral attacks. Purely by means of the Darwinian processes of inheritance, innovation and natural selection, bacteria found a way to cut little snippets of the viral DNA or RNA code from the viral invaders and store those snippets in its own loop of bacterial DNA. Through a complex process, these viral signatures are then used to find similar sections of viral DNA or RNA that might have found their way into a bacterial cell that is under attack by a virus. If such invading DNA or RNA is discovered by the bacterial antivirus CRISPR software, the CRISPR software will cut the invading DNA or RNA at that point. That is like chopping a virus in half with a guillotine. The virus then stops dead in its tracks. Biologists now use this bacterial CRISPR antiviral system to edit the DNA of the complex eukaryotic cells of higher life forms. They do this by building CRISPR-Cas9 complexes with guide RNA that matches the section of DNA that they wish to edit as shown in Figure 2. The CRISPR-Cas9 complex will then cut the target DNA at the specified point and a new section of DNA can then be inserted.
The antiviral software running on your hardware does the very same thing. The vendor of your antiviral software periodically updates a database on your hardware with new viral signatures that are snippets of virus code from the computer viruses that are running around the world. The antiviral software then scans your hardware for the viral signatures and if it finds any, the antiviral software destroys the viral infection. This is an example of convergence. In biology, convergence is the idea that sometimes organisms that are not at all related will come up with very similar solutions to common problems that they share. For example, the concept of the eye has independently evolved at least 40 different times in the past 600 million years, and there are many examples of “living fossils” showing the evolutionary path. For example, the camera-like structures of the human eye and the eye of an octopus are nearly identical, even though each structure evolved totally independent of the other.
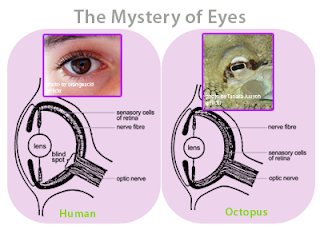
Figure 1 - The eye of a human and the eye of an octopus are nearly identical in structure, but evolved totally independently of each other. As Daniel Dennett pointed out, there are only a certain number of Good Tricks in Design Space and natural selection will drive different lines of descent towards them.
A good Wikipedia article on CRISPR can be found at:
CRISPR gene editing
https://en.wikipedia.org/wiki/CRISPR_gene_editing
Below is a very good YouTube video that quickly explains CRISPR in seven minutes and twenty seconds:
What is CRISPR?
https://www.youtube.com/watch?v=MnYppmstxIs
For a deeper dive, try this two-part YouTube video:
The principle of CRISPR System and CRISPR-CAS9 Technique (Part 1)
https://www.youtube.com/watch?v=VtOZdThI6dM
The principle of CRISPR System and CRISPR-CAS9 Technique (Part 2)
https://www.youtube.com/watch?v=KIMsVSQGBqw
Figure 2 – CRISPR-Cas9 acts like a line editor that allows biologists to locate a specific line of code in DNA and replace it with new DNA code.
The Major Advances in Working on Computer Source Code
Now let's look at the major developments that we have seen in writing and editing computer source code over the years to see where biological programming might be heading. Basically, we have had five major advances:
1. Punch Cards in the 1950s and 1960s
2. Line Editors like ed and ex on Unix in the early 1970s
3. The development of full-screen editors like ISPF on IBM mainframes and vi on Unix in the late 1970s
4. The development of IDEs like Eclipse or Microsoft's Visual Studio in the 1990s
5. The development of AI code generators in the 2020s
Now, I may be jumping the gun with the idea that AI code generators will appear in the 2020s, but we must take note that Microsoft has purchased the exclusive rights to the GPT-3 AI text generator that can now spit out computer source code with some acceptable level of bugs in it. For more on that see:
The Impact of GPT-3 AI Text Generation on the Development and Maintenance of Computer Software. But let's first go back to the 1950s and 1960s when we wrote computer source code on punch cards and see how the development of line editors and full-screen editors then improved programming efficiency by a factor of more than 100 times.
Programming on Punch Cards
Like the biological labwork of today, writing and maintaining software in the 20th century was very inefficient, time-consuming and tedious. For example, when I first learned to write Fortran code at the University of Illinois at Urbana in 1972, we were punching out programs on an IBM 029 keypunch machine, and I discovered that writing code on an IBM 029 keypunch machine was even worse than writing term papers on a manual typewriter. At least when you submitted a term paper with a few typos, your professor was usually kind enough not to abend your term paper right on the spot and give you a grade of zero. Sadly, I learned that such was not the case with Fortran compilers! The first thing you did was to write out your code on a piece of paper as best you could back at the dorm. The back of a large stack of fan-folded printer paper output was ideal for such purposes. In fact, as a physics major, I first got hooked by software while digging through the wastebaskets of DCL, the Digital Computing Lab, at the University of Illinois looking for fan-folded listings of computer dumps that were about a foot thick. I had found that the backs of thick computer dumps were ideal for working on lengthy problems in my quantum mechanics classes.
It paid to do a lot of desk-checking of your code back at the dorm before heading out to the DCL. Once you got to the DCL, you had to wait your turn for the next available IBM 029 keypunch machine. This was very much like waiting for the next available washing machine on a crowded Saturday morning at a laundromat. When you finally got to your IBM 029 keypunch machine, you would load it up with a deck of blank punch cards and then start punching out your program. You would first press the feed button to have the machine pull your first card from the deck of blank cards and register the card in the machine. Fortran compilers required code to begin in column 7 of the punch card so the first thing you did was to press the spacebar 6 times to get to column 7 of the card. Then you would try to punch in the first line of your code. If you goofed and hit the wrong key by accident while punching the card, you had to eject the bad card and start all over again with a new card. Structured programming had not been invented yet, so nobody indented code at the time. Besides, trying to remember how many times to press the spacebar for each new card in a block of indented code was just not practical. Pressing the spacebar 6 times for each new card was hard enough! Also, most times we proofread our card decks by flipping through them before we submitted the card deck. Trying to proofread indented code in a card deck would have been rather disorienting, so nobody even thought of indenting code. Punching up lots of comment cards was also a pain, so most people got by with a minimum of comment cards in their program deck.
After you punched up your program on a card deck, you would then punch up your data cards. Disk drives and tape drives did exist in those days, but disk drive storage was incredibly expensive and tapes were only used for huge amounts of data. If you had a huge amount of data, it made sense to put it on a tape because if you had several feet of data on cards, there was a good chance that the operator might drop your data card deck while feeding it into the card reader. But usually, you ended up with a card deck that held the source code for your program and cards for the data to be processed too. You also punched up the JCL (Job Control Language) cards for the IBM mainframe that instructed the IBM mainframe to compile, link and then run your program all in one run. You then dropped your finalized card deck into the input bin so that the mainframe operator could load your card deck into the card reader for the IBM mainframe. After a few hours, you would then return to the output room of the DCL and go to the alphabetically sorted output bins that held all the jobs that had recently run. If you were lucky, in your output bin you found your card deck and the fan-folded computer printout of your last run. Unfortunately, normally you found that something probably went wrong with your job. Most likely you had a typo in your code that had to be fixed. If it was nighttime and the mistake in your code was an obvious typo, you probably still had time for another run, so you would get back in line for an IBM 029 keypunch machine and start all over again. You could then hang around the DCL working on the latest round of problems in your quantum mechanics course. However, machine time was incredibly expensive in those days and you had a very limited budget for machine charges. So if there was some kind of logical error in your code, many times you had to head back to the dorm for some more desk checking of your code before giving it another shot the next day.

Figure 3 - An IBM 029 keypunch machine like the one I first learned to program on at the University of Illinois in 1972.

Figure 4 - Each card could hold a maximum of 80 bytes. Normally, one line of code was punched onto each card.

Figure 5 - The cards for a program were held together into a deck with a rubber band, or for very large programs, the deck was held in a special cardboard box that originally housed blank cards. Many times the data cards for a run followed the cards containing the source code for a program. The program was compiled and linked in two steps of the run and then the generated executable file processed the data cards that followed in the deck.

Figure 6 - To run a job, the cards in a deck were fed into a card reader, as shown on the left above, to be compiled, linked, and executed by a million-dollar mainframe computer with a clock speed of about 750 kHz and about 1 MB of memory.
Figure 7 - The output of a run was printed on fan-folded paper and placed into an output bin along with your input card deck.
I finished up my B.S. in Physics at the University of Illinois at Urbana in 1973 and headed up north to complete an M.S. in Geophysics at the University of Wisconsin at Madison. Then from 1975 – 1979, I was an exploration geophysicist exploring for oil, first with Shell, and then with Amoco. I kept coding Fortran the whole time. In 1979, I made a career change into IT and spent about 20 years in development. For the last 17 years of my career, I was in IT operations, supporting middleware on WebSphere, JBoss, Tomcat, and ColdFusion. In 1979, when I became an IT professional in Amoco's IT department, I noticed that not much had changed with the way software was developed and maintained. Structured programming had arrived, so we were now indenting code and adding comment statements to the code, but I was still programming on cards. We were now using IBM 129 keypunch machines that were a little bit more sophisticated than the old IBM 029 keypunch machines. However, the coding process was still very much the same. I worked on code at my desk and still spent a lot of time desk checking the code. When I was ready for my next run, I would get into an elevator and travel down to the basement of the Amoco Building where the IBM mainframes were located. Then I would punch my cards on one of the many IBM 129 keypunch machines but this time with no waiting for a machine. After I submitted my deck, I would travel up 30 floors to my cubicle to work on something else. After a couple of hours, I would head down to the basement again to collect my job. On a good day, I could manage to get 4 runs in. But machine time was still incredibly expensive. If I had a $100,000 project, $25,000 went for programming time, $25,000 went to IT overhead like management and data management services costs, and a full $50,000 went to machine costs for compiles and test runs!
This may all sound very inefficient and tedious today, but it can be even worse. I used to talk to the old-timers about the good old days of IT. They told me that when the operators began their shift on an old-time 1950s vacuum tube computer, the first thing they did was to crank up the voltage on the vacuum tubes to burn out the tubes that were on their last legs. Then they would replace the burned-out tubes to start the day with a fresh machine.
Figure 8 – In the 1950s, the electrical relays of the very ancient computers were replaced with vacuum tubes that were also very large, used lots of electricity and generated lots of waste heat too, but the vacuum tubes were 100,000 times faster than relays.
They also explained that the machines were so slow that they spent all day processing production jobs. Emergency maintenance work to fix production bugs was allowed at night, but new development was limited to one compile and test run per week! They also told me about programming the plugboards of electromechanical Unit Record Processing machines back in the 1950s by physically rewiring the plugboards. The Unit Record Processing machines would then process hundreds of punch cards per minute by routing the punch cards from machine to machine in processing streams.
Figure 9 – In the 1950s Unit Record Processing machines like this card sorter were programmed by physically rewiring a plugboard.
Figure 10 – The plugboard for a Unit Record Processing machine.
Programming With a Line Editor
A line editor lets you find and work on one line of code at a time just as a CRISPR-Cas9 complex lets you find and edit
one specific line of DNA code in a living cell. Line editors were first developed for computer terminals like teletype machines that were a combination of a keyboard and a printer. You could type in operating system commands on the teletype keyboard and the operating system of the computer would print back responses on a roll of paper - see Figure 11. You could also edit files on the teletype using a line editor. The Unix line editor ed was developed by Ken Thompson in August 1969 on a DEC PDP-7 at AT&T Bell Labs. The ed line editor was rather difficult to use, so Bill Joy wrote the ex line editor in 1976 for Unix. The ex line editor is still used today in Unix shell scripts to edit files.
Figure 11 – Some graduate students huddled around a DEC PDP-8/e minicomputer. Notice the teletype machines in the foreground on the left that were used to input code and data into the machine and to print out results as well.
The following Wikipedia articles are a good introduction to line editors.
Line Editor
https://en.wikipedia.org/wiki/Line_editor
ed (text editor)
https://en.wikipedia.org/wiki/Ed_(text_editor)
ex (text editor)
https://en.wikipedia.org/wiki/Ex_(text_editor)
The article below is a good reference if you would like to play with the ex line editor on a Unix system or under Cygwin running on a PC.
Introducing the ex Editor
https://www.cs.ait.ac.th/~on/O/oreilly/unix/vi/ch05_01.htm
You can download a free copy of Cygwin from the Cygwin organization at:
Cygwin
https://www.cygwin.com/
Now let's play with the ex line editor under Cygwin on my Windows laptop. In Figure 12 we see a 17 line C++ program that asks a user for two numbers and then prints out the sum of two numbers. The ex line editor usually only prints out one line at a time, but I was able to get ex to print out the whole program by entering the command "1,17". I can also find the first occurrence of the string "cout" using the command "/cout". In a manner similar to the way CRISPR-Cas9 can edit a specific line of DNA code, I can then change "cout" to "pout" with the command "s/cout/pout/". Finally, I can list out the whole program again with a "1,17" command to the ex line editor.
Figure 12 – A 17 line C++ program that asks for two numbers and then prints out the sum. To list the whole program, I type "1,17" into the ex line editor.
Figure 13 – To find the first occurrence of the string "cout", I type "/cout" into the ex line editor
Figure 14 – To change the first "cout" string to "pout", I type in "s/cout/pout/ into the ex line editor.
Figure 15 – Now I list the whole program with the "1,17" command. Notice that the first "cout" has been mutated to "pout". Such a devastating mutation means that the C++ program will not even compile into an executable file that can run on my computer.
As I mentioned, I finished up my B.S. in Physics at the University of Illinois at Urbana in 1973 and headed up north to complete an M.S. in Geophysics at the University of Wisconsin at Madison. I was working with a team of graduate students who were collecting electromagnetic data in the field on a DEC PDP-8/e minicomputer - see Figure 11. The machine cost about $30,000 in 1973 (about $176,000 in 2020 dollars) and was about the size of a large side-by-side refrigerator. The machine had 32 KB of magnetic core memory, about 2 million times less memory than a modern 64 GB smartphone. This was my first experience with interactive computing and using a line editor to create and edit code. Previously, I had only written Fortran programs for batch processing on IBM mainframes using an IBM 029 keypunch machine and punch cards. I wrote BASIC programs on the DEC PDP-8/e minicomputer using a teletype machine and a built-in line editor. The teletype machine was also used to print out program runs. My programs were saved to a magnetic tape and I could also read and write data from a tape as well. I found that using the built-in line editor made programming at least 10 times easier than writing code on punch cards. Now with a simple one-line command I could print out large portions of my code and see all of the code at the same time. When programming on cards, you could only look at code one card at a time. Now, whenever you made a run with a compile-link-run job on cards, you did get a listing of the complete program. But otherwise, you spent most of your time shuffling through a large deck of cards. That made it very difficult to find specific lines of code in your deck of cards or all the occurrences of a particular variable.
Another neat thing about the DEC PDP-8/e minicomputer was there were no computer charges and I did not have to wait hours to see the output of a run. None of my departmental professors knew how to program computers, so there was plenty of free machine time because only about four graduate students knew how to program at the time. I also learned the time-saving trick of interactive programming. Originally, I would write a BASIC program and hard-code the data values for a run directly in the code and then run the program as a batch job. After the run, I would then edit the code to change the hard-coded data values and then run the program again. Then one of my fellow graduate students showed me the trick of how to add a very primitive interactive user interface to my BASIC programs. Instead of hard-coding data values, my BASIC code would now prompt me for values that I could enter on the fly on the teletype machine. This allowed me to create a library of "canned" BASIC programs on tape that I never had to change. I could just run my "canned" programs with new input data as needed.
Programming With a Full-Screen Editor
In 1972, Dahl, Dijkstra, and Hoare published Structured Programming, in which they suggested that computer programs should have a complex internal structure with no GOTO statements, lots of subroutines, indented code, and many comment statements. But writing such complicated structures on punch cards was nearly impossible. Even with a line editor, it was very difficult to properly do all of the indentation and spacing. As I mentioned above, for several decades programmers writing code on punch cards hardly ever even got to see their code all in one spot on a listing. Mainly, we were rifling through card decks to find things. Trying to punch up structured code on punch cards would have just made a very difficult process much harder and trying to find things in a deck of structured code would have been even slower if there were lots of punch cards with just comment statements on them. So nobody really wrote structured code with punch cards or even a line editor. In response, in 1974 IBM introduced the SPF (Structured Programming Facility) full-screen editor. At the time, IBM was pushing SPF as a way to do the complicated coding that structured programming called for. Later, IBM renamed SPF to ISPF (Interactive Systems Productivity Facility).
Figure 16 - IBM 3278 terminals were connected to controllers that connected to IBM mainframes. The IBM 3278 terminals then ran interactive TSO sessions with the IBM mainframes. The ISPF full-screen editor was then brought up under TSO after you logged into a TSO session.
A full-screen editor like ISPF is much like a wordprocessing editor like Microsoft's Word or WordPad that displays a whole screen of code at one time. With your cursor, you can move to different lines of code and edit them directly by typing over old code. You can also insert new code and do global changes to the code. ISPF is still used today for IBM's z/OS and z/VM mainframe operating systems. In addition to a powerful full-screen editor, ISPF lets programmers run many utilities from a hierarchical set of menus rather than run them as TSO command-line commands. At first, in the late 1970s, the IBM 3278 terminals appeared in IT departments in "tube rows" like the IBM 029 keypunch machines of yore because machine costs for interactive TSO sessions were still much higher than the rate for batch jobs running off punch cards. Today, programmers do not worry much about machine costs, but in the early 1980s the machine costs for developing a new application still came to 50% of the entire budget for the project. But by the early 1980s, each IT professional was finally given their own IBM 3278 terminal on their own desk. Finally, there was no more waiting in line for an input device!
At the same time, the universities around the world were also building Computer Science Departments in the late 1960s and early 1970s. The universities found it much cheaper to teach students how to program on cheap Unix machines rather than IBM mainframes. So most universities became Unix shops teaching the C programming language. Now, when those students hit the corporate IT departments in the early 1980s, they brought along their fondness for the Unix operating system and the C programming language. But since most corporate IT departments still remained IBM shops, these students had to learn the IBM way of life. Still, the slow influx of programmers with experience with Unix and C running on cheap Unix servers eventually led to the Distributed Computing mass extinction event in the early 1990s. The Distributed Computing mass extinction event was much like the End-Permian greenhouse gas mass extinction 252 million years ago that killed off 95% of marine species and 70% of land-based species. The Distributed Computing mass extinction event ended the reign of the mainframes and replaced them with large numbers of cheap Unix servers in server farms. This greatly advanced the presence of Unix in IT shops around the world and the need for business programmers to learn Unix and a full-screen editor for Unix files. For more on that see Cloud Computing and the Coming Software Mass Extinction. Fortunately, Unix also had a very popular full-screen editor called vi. When Bill Joy wrote the Unix ex line editor in 1976, he also included a visual interface to it called "vi" that was sort of a cross between a line editor and a full-screen editor. The vi full-screen editor takes a bit of time to get used to, but once you get used to vi, it turns out to be a very powerful full-screen editor that is still used today by a large number of Unix programmers. In fact, I used vi to do all of my Unix editing until the day I retired in December of 2016. One of the advantages of using vi on Unix is that you can use a very similar Windows full-screen editor called Vim to edit Windows files. That way, your editing motor-memory works on both the Unix and Windows operating systems. Below is a good Wikipedia article on the vi editor.
vi
https://en.wikipedia.org/wiki/Vi
You can download the Vim editor at:
Vim
https://www.vim.org/
Figure 17 – Now take a look at our C++ program in the vi editor that is running under Cygwin on my Windows laptop. Notice that my cursor is now located at the end of line 17 after I easily added some comment lines and repeated them with the copy/paste function of the vi full-screen editor.
One of the other characteristics of structured programming was the use of "top-down" programming, where programs began execution in a mainline routine that then called many other subroutines. The purpose of the mainline routine was to perform simple high-level logic that called the subordinate subroutines in a fashion that was easy to follow. This structured programming technique made it much easier to maintain and enhance software by simply calling subroutines from the mainline routine in the logical manner required to perform the needed tasks, like assembling Lego building blocks into different patterns that produced an overall new structure. The structured approach to software also made it much easier to reuse software. All that was needed was to create a subroutine library of reusable source code that was already compiled. A mainline program that made calls to the subroutines of the subroutine library was compiled as before. The machine code for the previously compiled subroutines was then added to the resulting executable file by a linkage editor. This made it much easier for structured eukaryotic programs to use reusable code by simply putting the software "conserved core processes" into already compiled subroutine libraries. The vi full-screen editor running under Unix and the ISPF full-screen editor running under TSO on IBM 3278 terminals also made it much easier to reuse source code because now many lines of source code could be simply copied from one program file to another, with the files stored on disk drives, rather than on punch cards or magnetic tape.
Thus, not only did the arrival of full-screen editors make writing and maintaining code much easier, but they also ushered in a whole new way of coding. Now programmers were able to write structured code that was easier to support and maintain. They could also benefit from code reuse by being able to do easy copy/paste operations between files of source code and also by building libraries of reusable code that could be brought in at link-time. With a large library of reusable code, programmers could just write mainline programs that made calls to the subroutines in reusable code libraries. Complex living things do the same thing.
In Facilitated Variation and the Utilization of Reusable Code by Carbon-Based Life, I showcased the theory of facilitated variation by Marc W. Kirschner and John C. Gerhart. In The Plausibility of Life (2005), Marc W. Kirschner and John C. Gerhart present their theory of facilitated variation. The theory of facilitated variation maintains that, although the concepts and mechanisms of Darwin's natural selection are well understood, the mechanisms that brought forth viable biological innovations in the past are a bit wanting in classical Darwinian thought. In classical Darwinian thought, it is proposed that random genetic changes, brought on by random mutations to DNA sequences, can very infrequently cause small incremental enhancements to the survivability of the individual, and thus provide natural selection with something of value to promote in the general gene pool of a species. Again, as frequently cited, most random genetic mutations are either totally inconsequential, or totally fatal in nature, and consequently, are either totally irrelevant to the gene pool of a species or are quickly removed from the gene pool at best. The theory of facilitated variation, like classical Darwinian thought, maintains that the phenotype of an individual is key, and not so much its genotype since natural selection can only operate upon phenotypes. The theory explains that the phenotype of an individual is determined by a number of "constrained" and "deconstrained" elements. The constrained elements are called the "conserved core processes" of living things that essentially remain unchanged for billions of years, and which are to be found to be used by all living things to sustain the fundamental functions of carbon-based life, like the generation of proteins by processing the information that is to be found in DNA sequences, and processing it with mRNA, tRNA and ribosomes, or the metabolism of carbohydrates via the Krebs cycle. The deconstrained elements are weakly-linked regulatory processes that can change the amount, location and timing of gene expression within a body, and which, therefore, can easily control which conserved core processes are to be run by a cell and when those conserved core processes are to be run by them. The theory of facilitated variation maintains that most favorable biological innovations arise from minor mutations to the deconstrained weakly-linked regulatory processes that control the conserved core processes of life, rather than from random mutations of the genotype of an individual in general that would change the phenotype of an individual in a purely random direction. That is because the most likely change of direction for the phenotype of an individual, undergoing a random mutation to its genotype, is the death of the individual.
Marc W. Kirschner and John C. Gerhart begin by presenting the fact that simple prokaryotic bacteria, like E. coli, require a full 4,600 genes just to sustain the most rudimentary form of bacterial life, while much more complex multicellular organisms, like human beings, consisting of tens of trillions of cells differentiated into hundreds of differing cell types in the numerous complex organs of a body, require only a mere 22,500 genes to construct. The baffling question is, how is it possible to construct a human being with just under five times the number of genes as a simple single-celled E. coli bacterium? The authors contend that it is only possible for carbon-based life to do so by heavily relying upon reusable code in the genome of complex forms of carbon-based life.
Figure 18 – A simple single-celled E. coli bacterium is constructed using a full 4,600 genes.
Figure 19 – However, a human being, consisting of about 100 trillion cells that are differentiated into the hundreds of differing cell types used to form the organs of the human body, uses a mere 22,500 genes to construct a very complex body, which is just slightly under five times the number of genes used by simple E. coli bacteria to construct a single cell. How is it possible to explain this huge dynamic range of carbon-based life? Marc W. Kirschner and John C. Gerhart maintain that, like complex software, carbon-based life must heavily rely on the microservices of reusable code.
The authors then propose that complex living things arise by writing "deconstrained" mainline programs that then call the "constrained" libraries of reusable code that have been in use for billions of years.
Programming with an IDE (Integrated Development Environment)
IDEs first appeared in the early 1990s. An IDE assists developers with writing new software or maintaining old software by automating many of the labor-intensive and tedious chores of working on software. The chief advance that came with the early IDEs was their ability to do interactive full-screen debugging. With interactive full-screen debugging, programmers were able to step through their code line-by-line as the code actually executed. Programmers could set a breakpoint in their code, and when the breakpoint was reached as their code executed, the code stopped executing and put them into debug mode at the breakpoint. They could then look at the values of all of the variables used by their code. They could also step through the code line by line and watch the logical flow of their code operate as it branched around their program mainline and into and back from called functions. They could also watch the values of variables change as each new line of code ran. This provided a huge improvement in debugging productivity. Writing code is easy. Debugging code so that it actually works takes up about 95% of a programmer's time. Prior to full-screen debugging in an IDE, programmers would place a number of PRINT statements into their code for debugging purposes. For example, they might add the following line of code to their program: PRINT("This is breakpoint #1, TotalOrderItems, TotalOrderAmount). They would then run the program that they were debugging. After their program crashed, they would look at the output. If they saw something like "This is breakpoint #1 6 $4824.94", they would know that their code at least got to the debug PRINT statement before it crashed, and also that at that time, 6 items had been ordered for a total of $4824.94. By placing many PRINT debug statements in their code, programmers were able to very slowly identify all of the bugs in their program and then fix them.
Another major advance that IDEs brought was automatic code completion. This was especially important for object-oriented languages like C++ and Java (1995). With automatic code completion, the IDE full-screen editor would sense new code that was being written and would provide a drop-down of possible ways to complete the code being typed. For example, if the programmer typed in the name of an existing object the automatic code completion would display the list of methods defined by the object and the data attributes defined by the object. The programmer could then select the proper element from the drop-down and the code would then be completed by the IDE. My guess would be that the next IDE advance would be an AI form of super-code completion. For example, the GPT-3 AI text generator can now spit out computer source code with some acceptable level of bugs in it. If GPT-3 were allowed to read tons of software code for particular languages, it could get even better. For more on that see:
The Impact of GPT-3 AI Text Generation on the Development and Maintenance of Computer Software.
Microsoft's very first IDE was Visual C++ and came out in 1993. After developing several similar IDEs for different computer languages, Microsoft combined them all under a product called Visual Studio in 1997. Similarly, IBM released its Visual Age IDE in 1993. In 2004, Visual Age evolved into Eclipse 3.0 and was further developed and maintained by the open-source Eclipse Foundation. For those of you not familiar with Microsoft's Visual Studio IDE, you can read about it and download a free trial version of Visual Studio for your own personal use at:
Visual Studio
https://visualstudio.microsoft.com/vs/
If you do not need all of the bells and whistles of a modern IDE, you can download a permanent free copy of Visual Studio Code. Visual Studio Code is a very useful and free IDE with all of the powers of a 1990s-style IDE. It has interactive full-screen debugging and code completion and can be used for many different languages like C, C++, C# and Java.
Visual Studio Code
https://code.visualstudio.com/
When you crank up Microsoft's Visual Studio, you will find all of the software tools that you need to develop new software or maintain old software. The Visual Studio IDE allows software developers to perform development and maintenance chores that took days or weeks back in the 1960s, 1970s and 1980s in a matter of minutes.
Figure 20 – Above is a screenshot of Microsoft's Visual Studio IDE. It assists developers with writing new software or maintaining old software by automating many of the labor-intensive and tedious chores of working on software. Station B intends to do the same for biologists.
Before you download a free community version of Visual Studio be sure to watch some of the Visual Studio 2019 Launch videos at the bottom of the Visual Studio 2019 download webpage to get an appreciation for what a modern IDE can do.
You can also download a free permanent copy of the Eclipse IDE for many different computer languages at:
Eclipse Foundation
https://www.eclipse.org/
Figure 21 – A screenshot of the Eclipse IDE for C++.
Conclusion
As you can imagine, writing and maintaining modern software that might consist of millions of lines of code would have been very difficult back in the 1960s using punch cards. To address that problem, IT slowly came up with many software tools over the decades to help automate the process. Being able to quickly edit code and debug it was certainly a priority. But unfortunately, the progress of automating software development and maintenance was surprisingly very slow and was always impeded by the expediency of the moment. There always was the time-pressure of trying to get new software into Production. So rather than spending time to write automation software that could be used to improve the productivity of writing and maintaining software, IT Management always stressed the importance of hitting the next deadline with whatever incredibly inefficient technology we had at the time. When I first transitioned from being an exploration geophysicist to being an IT professional back in 1979, I was shocked by the very primitive development technology that IT had at hand. At the time, it seemed that IT Management wanted us to perform miracles on punch cards, sort of like painting the Eifel Tower with a single paintbrush and a bucket of paint in a single day. In response, in 1985 I developed my own mainframe-based IDE called BSDE (Bionic Systems Development Environment) at a time when IDEs did not exist. The BSDE IDE was used to "grow" applications in a biological manner from an "embryo" and from 1985 - 1992 BSDE was used to put several million lines of code into Production at Amoco. For more on BSDE see the last part of Programming Biology in the Biological Computation Group of Microsoft Research. Similarly, something like Microsoft's Station B initiative to build a biological IDE for writing and editing DNA code will surely be necessary to do advanced biological programing on the billions of DNA base pairs used by complex multicellular carbon-based life. That will certainly take a lot of time and money to accomplish, but I think that the long history of writing and maintaining computer software certainly shows that it will be well worth the while.
Comments are welcome at scj333@sbcglobal.net
To see all posts on softwarephysics in reverse order go to:
https://softwarephysics.blogspot.com/
Regards,
Steve Johnston















